Abstract
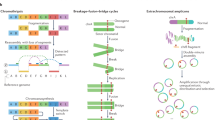
Structural and quantitative chromosomal rearrangements, collectively referred to as structural variation (SV), contribute to a large extent to the genetic diversity of the human genome and thus are of high relevance for cancer genetics, rare diseases and evolutionary genetics. Recent studies have shown that SVs can not only affect gene dosage but also modulate basic mechanisms of gene regulation. SVs can alter the copy number of regulatory elements or modify the 3D genome by disrupting higher-order chromatin organization such as topologically associating domains. As a result of these position effects, SVs can influence the expression of genes distant from the SV breakpoints, thereby causing disease. The impact of SVs on the 3D genome and on gene expression regulation has to be considered when interpreting the pathogenic potential of these variant types.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Sudmant, P. H. et al. An integrated map of structural variation in 2,504 human genomes. Nature 526, 75–81 (2015). This study provides a comprehensive structural variation map of >25,000 healthy human genomes.
Genome of the Netherlands Consortium. Whole-genome sequence variation, population structure and demographic history of the Dutch population. Nat. Genet. 46, 818–825 (2014).
Kloosterman, W. P. et al. Characteristics of de novo structural changes in the human genome. Genome Res. 25, 792–801 (2015).
Weischenfeldt, J., Symmons, O., Spitz, F. & Korbel, J. O. Phenotypic impact of genomic structural variation: insights from and for human disease. Nat. Rev. Genet. 14, 125–138 (2013).
Zarrei, M., MacDonald, J. R., Merico, D. & Scherer, S. W. A copy number variation map of the human genome. Nat. Rev. Genet. 16, 172–183 (2015).
Sebat, J. et al. Strong association of de novo copy number mutations with autism. Science 316, 445–449 (2007).
Soemedi, R. et al. Contribution of global rare copy-number variants to the risk of sporadic congenital heart disease. Am. J. Hum. Genet. 91, 489–501 (2012).
Xu, B. et al. Strong association of de novo copy number mutations with sporadic schizophrenia. Nat. Genet. 40, 880–885 (2008).
Walsh, T. et al. Rare structural variants disrupt multiple genes in neurodevelopmental pathways in schizophrenia. Science 320, 539–543 (2008).
Cooper, G. M. et al. A copy number variation morbidity map of developmental delay. Nat. Genet. 43, 838–846 (2011).
Yang, L. et al. Diverse mechanisms of somatic structural variations in human cancer genomes. Cell 153, 919–929 (2013).
de Leeuw, N. et al. Diagnostic interpretation of array data using public databases and internet sources. Hum. Mutat. 33, 930–940 (2012).
Biederman, B. & Bowen, P. Balanced translocations involving chromosome 12: report of a case and possible evidence for position effect. Ann. Genet. 19, 257–260 (1976).
Hecht, F. & Kaiser-McCaw, B. Position effect in 8;14 translocation in Burkitt’s lymphoma. N. Engl. J. Med. 304, 174–175 (1981).
Venter, J. C. et al. The sequence of the human genome. Science 291, 1304–1351 (2001).
Roadmap Epigenomics Consortium et al. Integrative analysis of 111 reference human epigenomes. Nature 518, 317–330 (2015).
The ENCODE Project Consortium. An integrated encyclopedia of DNA elements in the human genome. Nature 489, 57–74 (2012).
Carvalho, C. M. & Lupski, J. R. Mechanisms underlying structural variant formation in genomic disorders. Nat. Rev. Genet. 17, 224–238 (2016).
Pombo, A. & Dillon, N. Three-dimensional genome architecture: players and mechanisms. Nat. Rev. Mol. Cell Biol. 16, 245–257 (2015).
Bonev, B. & Cavalli, G. Organization and function of the 3D genome. Nat. Rev. Genet. 17, 772 (2016).
Krijger, P. H. & de Laat, W. Regulation of disease-associated gene expression in the 3D genome. Nat. Rev. Mol. Cell Biol. 17, 771–782 (2016).
Alkan, C., Coe, B. P. & Eichler, E. E. Genome structural variation discovery and genotyping. Nat. Rev. Genet. 12, 363–376 (2011).
Wright, C. F., FitzPatrick, D. R. & Firth, H. V. Paediatric genomics: diagnosing rare disease in children. Nat. Rev. Genet. 19 253–268 (2018).
Gilissen, C. et al. Genome sequencing identifies major causes of severe intellectual disability. Nature 511, 344–347 (2014). This landmark study uses whole-genome sequencing to identify complex de novo SV in families with intellectual disability, including single exon deletions and insertional duplications.
Cretu Stancu, M. et al. Mapping and phasing of structural variation in patient genomes using nanopore sequencing. Nat. Commun. 8, 1326 (2017).
Huddleston, J. et al. Discovery and genotyping of structural variation from long-read haploid genome sequence data. Genome Res. 27, 677–685 (2017).
Chaisson, M. J. P. et al. Multi-platform discovery of haplotype-resolved structural variation in human genomes. Preprint at bioRxiv https://doi.org/10.1101/193144 (2017).
de Vries, B. B. et al. Diagnostic genome profiling in mental retardation. Am. J. Hum. Genet. 77, 606–616 (2005).
Talkowski, M. E. et al. Sequencing chromosomal abnormalities reveals neurodevelopmental loci that confer risk across diagnostic boundaries. Cell 149, 525–537 (2012).
Marshall, C. R. et al. Structural variation of chromosomes in autism spectrum disorder. Am. J. Hum. Genet. 82, 477–488 (2008).
Flottmann, R. et al. Noncoding copy-number variations are associated with congenital limb malformation. Genet. Med. https://doi.org/10.1038/gim.2017.154 (2017).
Sanders, S. J. et al. Insights into autism spectrum disorder genomic architecture and biology from 71 risk loci. Neuron 87, 1215–1233 (2015).
The 1000 Genomes Project Consortium et al. A global reference for human genetic variation. Nature 526, 68–74 (2015).
Hehir-Kwa, J. Y. et al. A high-quality human reference panel reveals the complexity and distribution of genomic structural variants. Nat. Commun. 7, 12989 (2016).
Brandler, W. M. et al. Frequency and complexity of de novo structural mutation in autism. Am. J. Hum. Genet. 98, 667–679 (2016).
Goldmann, J. M. et al. Parent-of-origin-specific signatures of de novo mutations. Nat. Genet. 48, 935–939 (2016).
Lelieveld, S. H. et al. Spatial clustering of de novo missense mutations identifies candidate neurodevelopmental disorder-associated genes. Am. J. Hum. Genet. 101, 478–484 (2017).
Turner, T. N. et al. Genomic patterns of de novo mutation in simplex autism. Cell 171, 710–722.e12 (2017). This study combines six SV calling algorithms to identify all de novo SVs in 516 families with autism.
Turner, T. N. et al. Genome sequencing of autism-affected families reveals disruption of putative noncoding regulatory DNA. Am. J. Hum. Genet. 98, 58–74 (2016).
Brandler, W. M. et al. Paternally inherited noncoding structural variants contribute to autism. Preprint at bioRxiv https://doi.org/10.1101/102327 (2017).
Werling, D. M. et al. Limited contribution of rare, noncoding variation to autism spectrum disorder from sequencing of 2,076 genomes in quartet families. Preprint at bioRxiv https://doi.org/10.1101/127043 (2017).
Siva, N. UK gears up to decode 100,000 genomes from NHS patients. Lancet 385, 103–104 (2015).
Deciphering Developmental Disorders Study. Prevalence and architecture of de novo mutations in developmental disorders. Nature 542, 433–438 (2017).
MacDonald, J. R., Ziman, R., Yuen, R. K., Feuk, L. & Scherer, S. W. The Database of Genomic Variants: a curated collection of structural variation in the human genome. Nucleic Acids Res. 42, D986–D992 (2014).
van Bon, B. W. M., Mefford, H. C. & de Vries, B. B. A. 15q13.3 Microdeletion. GeneReviews https://www.ncbi.nlm.nih.gov/books/NBK50780/ (23 July 2015).
Klopocki, E. et al. Complex inheritance pattern resembling autosomal recessive inheritance involving a microdeletion in thrombocytopenia-absent radius syndrome. Am. J. Hum. Genet. 80, 232–240 (2007).
Lupski, J. R. et al. Gene dosage is a mechanism for Charcot-Marie-Tooth disease type 1A. Nat. Genet. 1, 29–33 (1992).
Lek, M. et al. Analysis of protein-coding genetic variation in 60,706 humans. Nature 536, 285–291 (2016).
Lindsay, E. A. et al. Tbx1 haploinsufficieny in the DiGeorge syndrome region causes aortic arch defects in mice. Nature 410, 97–101 (2001).
Merscher, S. et al. TBX1 is responsible for cardiovascular defects in velo-cardio-facial/DiGeorge syndrome. Cell 104, 619–629 (2001).
Zollino, M. et al. Mutations in KANSL1 cause the 17q21.31 microdeletion syndrome phenotype. Nat. Genet. 44, 636–638 (2012).
Talkowski, M. E. et al. Assessment of 2q23.1 microdeletion syndrome implicates MBD5 as a single causal locus of intellectual disability, epilepsy, and autism spectrum disorder. Am. J. Hum. Genet. 89, 551–563 (2011).
Slager, R. E., Newton, T. L., Vlangos, C. N., Finucane, B. & Elsea, S. H. Mutations in RAI1 associated with Smith-Magenis syndrome. Nat. Genet. 33, 466–468 (2003).
Lupski, J. R. et al. DNA duplication associated with Charcot-Marie-Tooth disease type 1A. Cell 66, 219–232 (1991).
Chance, P. F. et al. DNA deletion associated with hereditary neuropathy with liability to pressure palsies. Cell 72, 143–151 (1993).
Cremer, T. & Cremer, C. Chromosome territories, nuclear architecture and gene regulation in mammalian cells. Nat. Rev. Genet. 2, 292–301 (2001).
Saurin, A. J. et al. The human polycomb group complex associates with pericentromeric heterochromatin to form a novel nuclear domain. J. Cell Biol. 142, 887–898 (1998).
Wansink, D. G. et al. Fluorescent labeling of nascent RNA reveals transcription by RNA polymerase II in domains scattered throughout the nucleus. J. Cell Biol. 122, 283–293 (1993).
Lieberman-Aiden, E. et al. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science 326, 289–293 (2009). This study presents the original description of the HiC protocol to analyse genome-wide chromatin interactions.
Dekker, J., Marti-Renom, M. A. & Mirny, L. A. Exploring the three-dimensional organization of genomes: interpreting chromatin interaction data. Nat. Rev. Genet. 14, 390–403 (2013).
Rao, S. S. et al. A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell 159, 1665–1680 (2014).
Bianco, S. et al. Polymer physics predicts the effects of structural variants on chromatin architecture. Nat. Genet. https://doi.org/10.1038/s41588-018-0098-8 (2018).
Franke, M. et al. Formation of new chromatin domains determines pathogenicity of genomic duplications. Nature 538, 265–269 (2016). This study shows that duplications can result in the formation of new TADs, a process that determines the pathogenicity of these rearrangements.
Burton, J. N. et al. Chromosome-scale scaffolding of de novo genome assemblies based on chromatin interactions. Nat. Biotechnol. 31, 1119–1125 (2013).
Rickman, D. S. et al. Oncogene-mediated alterations in chromatin conformation. Proc. Natl Acad. Sci. USA 109, 9083–9088 (2012).
Harewood, L. et al. Hi-C as a tool for precise detection and characterisation of chromosomal rearrangements and copy number variation in human tumours. Genome Biol. 18, 125 (2017).
Montavon, T. et al. A regulatory archipelago controls Hox genes transcription in digits. Cell 147, 1132–1145 (2011).
Spitz, F. & Furlong, E. E. Transcription factors: from enhancer binding to developmental control. Nat. Rev. Genet. 13, 613–626 (2012).
Kagey, M. H. et al. Mediator and cohesin connect gene expression and chromatin architecture. Nature 467, 430–435 (2010).
Splinter, E. et al. CTCF mediates long-range chromatin looping and local histone modification in the beta-globin locus. Genes Dev. 20, 2349–2354 (2006).
Palstra, R. J. et al. The beta-globin nuclear compartment in development and erythroid differentiation. Nat. Genet. 35, 190–194 (2003).
de Wit, E. & de Laat, W. A decade of 3C technologies: insights into nuclear organization. Genes Dev. 26, 11–24 (2012).
Dekker, J., Rippe, K., Dekker, M. & Kleckner, N. Capturing chromosome conformation. Science 295, 1306–1311 (2002).
Dixon, J. R. et al. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature 485, 376–380 (2012).
Nora, E. P., Dekker, J. & Heard, E. Segmental folding of chromosomes: a basis for structural and regulatory chromosomal neighborhoods? Bioessays 35, 818–828 (2013).
Lupianez, D. G. et al. Disruptions of topological chromatin domains cause pathogenic rewiring of gene-enhancer interactions. Cell 161, 1012–1025 (2015). This study demonstrates that TAD fusion (deletions) and TAD shuffling (inversions) can cause congenital disease.
Ruf, S. et al. Large-scale analysis of the regulatory architecture of the mouse genome with a transposon-associated sensor. Nat. Genet. 43, 379–386 (2011).
Symmons, O. et al. Functional and topological characteristics of mammalian regulatory domains. Genome Res. 24, 390–400 (2014).
Marinic, M., Aktas, T., Ruf, S. & Spitz, F. An integrated holo-enhancer unit defines tissue and gene specificity of the Fgf8 regulatory landscape. Dev. Cell 24, 530–542 (2013).
Dixon, J. R., Gorkin, D. U. & Ren, B. Chromatin domains: the unit of chromosome organization. Mol. Cell 62, 668–680 (2016).
Noordermeer, D. et al. Variegated gene expression caused by cell-specific long-range DNA interactions. Nat. Cell Biol. 13, 944–951 (2011).
Symmons, O. et al. The Shh topological domain facilitates the action of remote enhancers by reducing the effects of genomic distances. Dev. Cell 39, 529–543 (2016).
Lettice, L. A. et al. Opposing functions of the ETS factor family define Shh spatial expression in limb buds and underlie polydactyly. Dev. Cell 22, 459–467 (2012).
Kvon, E. Z. et al. Progressive Loss of function in a limb enhancer during snake evolution. Cell 167, 633–642.e11 (2016).
Moorthy, S. D. et al. Enhancers and super-enhancers have an equivalent regulatory role in embryonic stem cells through regulation of single or multiple genes. Genome Res. 27, 246–258 (2017).
Cannavo, E. et al. Shadow enhancers are pervasive features of developmental regulatory networks. Curr. Biol. 26, 38–51 (2016).
Hnisz, D. et al. Super-enhancers in the control of cell identity and disease. Cell 155, 934–947 (2013).
Long, H. K., Prescott, S. L. & Wysocka, J. Ever-changing landscapes: transcriptional enhancers in development and evolution. Cell 167, 1170–1187 (2016).
Will, A. J. et al. Composition and dosage of a multipartite enhancer cluster control developmental expression of Ihh (Indian hedgehog). Nat. Genet. 49, 1539–1545 (2017).
St-Jacques, B., Hammerschmidt, M. & McMahon, A. P. Indian hedgehog signaling regulates proliferation and differentiation of chondrocytes and is essential for bone formation. Genes Dev. 13, 2072–2086 (1999).
Hay, D. et al. Genetic dissection of the alpha-globin super-enhancer in vivo. Nat. Genet. 48, 895–903 (2016).
Shin, H. Y. et al. Hierarchy within the mammary STAT5-driven Wap super-enhancer. Nat. Genet. 48, 904–911 (2016).
Fulco, C. P. et al. Systematic mapping of functional enhancer-promoter connections with CRISPR interference. Science 354, 769–773 (2016).
Dickel, D. E. et al. Ultraconserved enhancers are required for normal development. Cell 172, 491–499.e15 (2018).
Osterwalder, M. et al. Enhancer redundancy provides phenotypic robustness in mammalian development. Nature 554, 239–243 (2018).
Zabidi, M. A. et al. Enhancer-core-promoter specificity separates developmental and housekeeping gene regulation. Nature 518, 556–559 (2015).
Lohan, S. et al. Microduplications encompassing the Sonic hedgehog limb enhancer ZRS are associated with Haas-type polysyndactyly and Laurin-Sandrow syndrome. Clin. Genet. 86, 318–325 (2014).
Klopocki, E. et al. A microduplication of the long range SHH limb regulator (ZRS) is associated with triphalangeal thumb-polysyndactyly syndrome. J. Med. Genet. 45, 370–375 (2008).
Benko, S. et al. Highly conserved non-coding elements on either side of SOX9 associated with Pierre Robin sequence. Nat. Genet. 41, 359–364 (2009).
Nora, E. P. et al. Spatial partitioning of the regulatory landscape of the X-inactivation centre. Nature 485, 381–385 (2012). References 74 and 100 demonstrate the presence of TADs in the mammalian genome.
Spielmann, M. & Mundlos, S. Structural variations, the regulatory landscape of the genome and their alteration in human disease. Bioessays 35, 533–543 (2013).
Spielmann, M. et al. Homeotic arm-to-leg transformation associated with genomic rearrangements at the PITX1 locus. Am. J. Hum. Genet. 91, 629–635 (2012).
Birnbaum, R. Y. et al. Coding exons function as tissue-specific enhancers of nearby genes. Genome Res. 22, 1059–1068 (2012).
Tayebi, N. et al. Deletions of exons with regulatory activity at the DYNC1I1 locus are associated with split-hand/split-foot malformation: array CGH screening of 134 unrelated families. Orphanet J. Rare Dis. 9, 108 (2014).
Lango Allen, H. et al. Next generation sequencing of chromosomal rearrangements in patients with split-hand/split-foot malformation provides evidence for DYNC1I1 exonic enhancers of DLX5/6 expression in humans. J. Med. Genet. 51, 264–267 (2014).
Ghiasvand, N. M. et al. Deletion of a remote enhancer near ATOH7 disrupts retinal neurogenesis, causing NCRNA disease. Nat. Neurosci. 14, 578–586 (2011).
Bhatia, S. et al. Disruption of autoregulatory feedback by a mutation in a remote, ultraconserved PAX6 enhancer causes aniridia. Am. J. Hum. Genet. 93, 1126–1134 (2013).
Dathe, K. et al. Duplications involving a conserved regulatory element downstream of BMP2 are associated with brachydactyly type A2. Am. J. Hum. Genet. 84, 483–492 (2009).
Sun, M. et al. Triphalangeal thumb-polysyndactyly syndrome and syndactyly type IV are caused by genomic duplications involving the long range, limb-specific SHH enhancer. J. Med. Genet. 45, 589–595 (2008).
Klopocki, E. et al. Copy-number variations involving the IHH locus are associated with syndactyly and craniosynostosis. Am. J. Hum. Genet. 88, 70–75 (2011).
Ngcungcu, T. et al. Duplicated enhancer region increases expression of CTSB and segregates with Keratolytic winter erythema in South African and Norwegian families. Am. J. Hum. Genet. 100, 737–750 (2017).
Cox, J. J., Willatt, L., Homfray, T. & Woods, C. G. A. SOX9 duplication and familial 46,XX developmental testicular disorder. N. Engl. J. Med. 364, 91–93 (2011).
Lupianez, D. G., Spielmann, M. & Mundlos, S. Breaking TADs: how alterations of chromatin domains result in disease. Trends Genet. 32, 225–237 (2016).
Lettice, L. A. et al. Enhancer-adoption as a mechanism of human developmental disease. Hum. Mutat. 32, 1492–1499 (2011). This study describes the disease mechanism of enhancer adoption in developmental disease.
Northcott, P. A. et al. Enhancer hijacking activates GFI1 family oncogenes in medulloblastoma. Nature 511, 428–434 (2014). This study reports the phenomenon of enhancer hijacking in cancer.
Redin, C. et al. The genomic landscape of balanced cytogenetic abnormalities associated with human congenital anomalies. Nat. Genet. 49, 36–45 (2017). By mapping the breakpoints of 273 balanced cytogenetic abnormalities, this study shows that TAD disruption is frequently associated with human congenital anomalies.
Northcott, P. A. et al. The whole-genome landscape of medulloblastoma subtypes. Nature 547, 311–317 (2017). This study shows that TAD shuffling and enhancer hijacking are frequent causes of medulloblastoma, a highly malignant childhood brain tumour.
Ordulu, Z. et al. Structural chromosomal rearrangements require nucleotide-level resolution: lessons from next-generation sequencing in prenatal diagnosis. Am. J. Hum. Genet. 99, 1015–1033 (2016).
Pfeifer, D. et al. Campomelic dysplasia translocation breakpoints are scattered over 1 Mb proximal to SOX9: evidence for an extended control region. Am. J. Hum. Genet. 65, 111–124 (1999).
Hnisz, D. et al. Activation of proto-oncogenes by disruption of chromosome neighborhoods. Science 351, 1454–1458 (2016).
Weischenfeldt, J. et al. Pan-cancer analysis of somatic copy-number alterations implicates IRS4 and IGF2 in enhancer hijacking. Nat. Genet. 49, 65–74 (2017). This paper describes a computational framework termed cis-expression structural alteration mapping (CESAM) to detect cancer-related gene overexpression resulting from TAD reorganization.
Kurth, I. et al. Duplications of noncoding elements 5’ of SOX9 are associated with brachydactyly-anonychia. Nat. Genet. 41, 862–863 (2009).
Zepeda-Mendoza, C. J. et al. Computational prediction of position effects of apparently balanced human chromosomal rearrangements. Am. J. Hum. Genet. 101, 206–217 (2017).
Ibn-Salem, J. et al. Deletions of chromosomal regulatory boundaries are associated with congenital disease. Genome Biol. 15, 423 (2014).
Sanchis-Juan, A. et al. Complex structural variants resolved by short-read and long-read whole genome sequencing in mendelian disorders. Preprint at bioRxiv https://doi.org/10.1101/281683 (2018).
Smedley, D. et al. A whole-genome analysis framework for effective identification of pathogenic regulatory variants in Mendelian disease. Am. J. Hum. Genet. 99, 595–606 (2016).
Duan, Z. et al. A three-dimensional model of the yeast genome. Nature 465, 363–367 (2010).
Chiariello, A. M., Annunziatella, C., Bianco, S., Esposito, A. & Nicodemi, M. Polymer physics of chromosome large-scale 3D organisation. Sci. Rep. 6, 29775 (2016).
Giorgetti, L. et al. Predictive polymer modeling reveals coupled fluctuations in chromosome conformation and transcription. Cell 157, 950–963 (2014).
Sanborn, A. L. et al. Chromatin extrusion explains key features of loop and domain formation in wild-type and engineered genomes. Proc. Natl Acad. Sci. USA 112, E6456–6465 (2015).
Fudenberg, G. et al. Formation of chromosomal domains by loop extrusion. Cell Rep. 15, 2038–2049 (2016).
Sexton, T. et al. Three-dimensional folding and functional organization principles of the Drosophila genome. Cell 148, 458–472 (2012).
Phillips-Cremins, J. E. et al. Architectural protein subclasses shape 3D organization of genomes during lineage commitment. Cell 153, 1281–1295 (2013).
Andrey, G. et al. Characterization of hundreds of regulatory landscapes in developing limbs reveals two regimes of chromatin folding. Genome Res. 27, 223–233 (2017).
Nora, E. P. et al. Targeted degradation of CTCF decouples local insulation of chromosome domains from genomic compartmentalization. Cell 169, 930–944.e22 (2017).
Schwarzer, W. et al. Two independent modes of chromatin organization revealed by cohesin removal. Nature 551, 51–56 (2017).
Benko, S. et al. Disruption of a long distance regulatory region upstream of SOX9 in isolated disorders of sex development. J. Med. Genet. 48, 825–830 (2011).
Chapuy, B. et al. Discovery and characterization of super-enhancer-associated dependencies in diffuse large B cell lymphoma. Cancer Cell 24, 777–790 (2013).
Giorgio, E. et al. A large genomic deletion leads to enhancer adoption by the lamin B1 gene: a second path to autosomal dominant adult-onset demyelinating leukodystrophy (ADLD). Hum. Mol. Genet. 24, 3143–3154 (2015).
Flottmann, R. et al. Microdeletions on 6p22.3 are associated with mesomelic dysplasia Savarirayan type. J. Med. Genet. 52, 476–483 (2015).
Niedermaier, M. et al. An inversion involving the mouse Shh locus results in brachydactyly through dysregulation of Shh expression. J. Clin. Invest. 115, 900–909 (2005).
Shimojima, K. et al. De novo microdeletion of 5q14.3 excluding MEF2C in a patient with infantile spasms, microcephaly, and agenesis of the corpus callosum. Am. J. Med. Genet. Part A 158A, 2272–2276 (2012).
Goubau, C. et al. Platelet defects in congenital variant of Rett syndrome patients with FOXG1 mutations or reduced expression due to a position effect at 14q12. Eur. J. Hum. Genet. 21, 1349–1355 (2013).
Acknowledgements
The authors apologize to colleagues whose work we were unable to discuss or failed to cite owing to space constraints. We thank members of the Mundlos and the Shendure laboratories for helpful discussions. M.S. was supported by a grant from the Deutsche Forschungsgemeinschaft (SP1532/2-1). D.G.L. is supported by a grant from the Deutsche Forschungsgemeinschaft (GA2495/1-1). Work in S.M.’s laboratory is funded by the Deutsche Forschungsgemeinschaft, the Berlin Institute for Health and the Max Planck Foundation.
Author information
Authors and Affiliations
Contributions
All authors contributed equally to all aspects of the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
RELATED LINKS
Database of Genomic Variants: http://dgv.tcag.ca/
3D Genome Browser: http://promoter.bx.psu.edu/hi-c/
VISTA Enhancer Browser: https://enhancer.lbl.gov/
DECIPHER: https://decipher.sanger.ac.uk/
Glossary
- Structural variation
-
(SV). Genetic variation that includes all structural and quantitative chromosomal rearrangements, that is, deletions and duplications, as well as copy-number-neutral aberrations, such as inversions, insertions and translocations.
- Copy number variation
-
(CNV). Genetic variation that refers only to quantitative chromosomal rearrangements, such as deletions and duplications.
- Microdeletion and microduplication syndromes
-
A group of syndromes that are caused by chromosomal microdeletions and microduplications, which make up a subset of copy number variations that are usually smaller than 5 Mb. Classic examples include the 7q11 deletion (Williams–Beuren syndrome), the 15q11–15q13 deletion (Prader–Willi and Angelman syndromes) and the 17p11 deletion (Smith–Magenis syndrome).
- Position effects
-
Effects of structural variation (SV), classically translocations, on the expression of a gene without any changes to its coding sequence or promoter region. These effects can also be observed if a gene is inserted into different regions of the genome or if SVs connect previously unconnected genes and their regulatory units. In these cases, the change in the level of gene expression is thought to result from changes in the position of the gene relative to its normal non-coding cis-regulatory environment.
- Chromosome conformation capture
-
(3C). PCR-based proximity-ligation analysis method of 3D genome organization that allows a reconstruction of the native chromatin structure within the nucleus. 3C employs a PCR-based approach to confirm interactions between two previously known loci (one versus one). The introduction of a second round of digestion and ligation allows the generation of self-circularized DNA fragments (4C-seq), on which inverse PCR can be used to identify all unknown fragments interacting with a specific locus (one versus all). Multiple genomic regions can be investigated in parallel by a multiplexing approach (5C; many versus many). HiC allows the identification of every possible interaction occurring in the nucleus through the introduction of biotin labelling to pull down ligation junctions (all versus all).
- Penetrance
-
A measure of the proportion of people with a particular genetic change (such as a mutation in a specific gene) who exhibit signs and symptoms of a genetic disorder. If some people with the mutation do not develop features of the disorder, the condition is said to have reduced (or incomplete) penetrance.
- Gene dosage
-
The number of copies of a particular gene, including all its regulatory regions present in a genome.
- Haploinsufficiency
-
A state in which one copy of a gene is inactivated or deleted and expression of the remaining functional copy of the gene is not sufficient to preserve normal function.
- Topologically associating domains
-
(TADs). Genomic sequences in the range of megabases in length that are separated by boundary regions and that physically interact with themselves more frequently than with the rest of the genome.
- Intra-TAD SVs
-
Structural variation (SV) that occurs within topologically associating domains (TADs). Within a regulatory domain, SVs of regulatory elements can change the enhancer dosage and may result in a tissue-specific loss of function (deletion) or a gain of function (duplication) of their endogenous target gene, which can be located anywhere within the TAD.
- Enhancer adoption
-
A phenomenon that refers to a regulatory gain of function mutation in which ectopic expression of a gene is driven by an enhancer that normally regulates another gene located in a different regulatory domain; also known as enhancer hijacking.
- Inter-TAD SVs
-
Structural variations (SVs) that occur between topologically associating domains (TADs). Such SVs affect several regulatory domains and have the potential to disrupt and rearrange the complex 3D chromatin organization of a locus by repositioning TAD boundaries, genes and enhancer elements. These position effects may lead to TAD fusion (deletion), neo-TAD formation (duplications) or TAD shuffling (inversion and/or translocation).
- TAD shuffling
-
The reordering of topologically associating domains (TADs) within the genome. Inversions that cross TAD boundaries or translocations can result in the fusion of two regulatory domains that do not naturally belong together, causing enhancer adoption. At the same time, these structural variations can result in regulatory loss of function by removing enhancer elements from their target genes.
Rights and permissions
About this article
Cite this article
Spielmann, M., Lupiáñez, D.G. & Mundlos, S. Structural variation in the 3D genome. Nat Rev Genet 19, 453–467 (2018). https://doi.org/10.1038/s41576-018-0007-0
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41576-018-0007-0
This article is cited by
-
Subtelomeric plasticity contributes to gene family expansion in the human parasitic flatworm Schistosoma mansoni
BMC Genomics (2024)
-
Random walk with restart on multilayer networks: from node prioritisation to supervised link prediction and beyond
BMC Bioinformatics (2024)
-
Alteration of chromosome structure impacts gene expressions implicated in pancreatic ductal adenocarcinoma cells
BMC Genomics (2024)
-
Genetic variation across and within individuals
Nature Reviews Genetics (2024)
-
ZNF689 deficiency promotes intratumor heterogeneity and immunotherapy resistance in triple-negative breast cancer
Cell Research (2024)