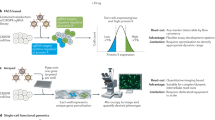
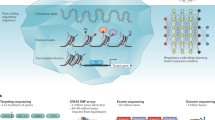
Abstract
We are entering a new era of mouse phenomics, driven by large-scale and economical generation of mouse mutants coupled with increasingly sophisticated and comprehensive phenotyping. These studies are generating large, multidimensional gene–phenotype data sets, which are shedding new light on the mammalian genome landscape and revealing many hitherto unknown features of mammalian gene function. Moreover, these phenome resources provide a wealth of disease models and can be integrated with human genomics data as a powerful approach for the interpretation of human genetic variation and its relationship to disease. In the future, the development of novel phenotyping platforms allied to improved computational approaches, including machine learning, for the analysis of phenotype data will continue to enhance our ability to develop a comprehensive and powerful model of mammalian gene–phenotype space.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Brown, S. D., Wurst, W., Kuhn, R. & Hancock, J. M. The functional annotation of mammalian genomes: the challenge of phenotyping. Annu. Rev. Genet. 43, 305–333 (2009).
Doyle, A., McGarry, M. P., Lee, N. A. & Lee, J. J. The construction of transgenic and gene knockout/knockin mouse models of human disease. Transgenic Res. 21, 327–349 (2012).
Bouabe, H. & Okkenhaug, K. Gene targeting in mice: a review. Methods Mol. Biol. 1064, 315–336 (2013).
Wang, H. et al. One-step generation of mice carrying mutations in multiple genes by CRISPR/Cas-mediated genome engineering. Cell 153, 910–918 (2013).
Fernandez, A., Josa, S. & Montoliu, L. A history of genome editing in mammals. Mamm. Genome 28, 237–246 (2017).
Birling, M. C., Herault, Y. & Pavlovic, G. Modeling human disease in rodents by CRISPR/Cas9 genome editing. Mamm. Genome 28, 291–301 (2017).
Hrabe de Angelis, M. H. et al. Genome-wide, large-scale production of mutant mice by ENU mutagenesis. Nat. Genet. 25, 444–447 (2000).
Nolan, P. M. et al. A systematic, genome-wide, phenotype-driven mutagenesis programme for gene function studies in the mouse. Nat. Genet. 25, 440–443 (2000). References 7 and 8 are instrumental in demonstrating the power of comprehensive phenotyping pipelines in large-scale mutagenesis screens.
Takeda, J., Keng, V. W. & Horie, K. Germline mutagenesis mediated by Sleeping Beauty transposon system in mice. Genome Biol. 8 (Suppl. 1), S14 (2007).
White, J. K. et al. Genome-wide generation and systematic phenotyping of knockout mice reveals new roles for many genes. Cell 154, 452–464 (2013).
de Angelis, M. H. et al. Analysis of mammalian gene function through broad-based phenotypic screens across a consortium of mouse clinics. Nat. Genet. 47, 969–978 (2015). References 10 and 11 describe the generation and phenotyping of hundreds of knockout mouse lines, revealing extensive pleiotropy and laying the groundwork for the IMPC.
Brown, S. D. & Moore, M. W. The International Mouse Phenotyping Consortium: past and future perspectives on mouse phenotyping. Mamm. Genome 23, 632–640 (2012).
Visscher, P. M. & Yang, J. A plethora of pleiotropy across complex traits. Nat. Genet. 48, 707–708 (2016).
Dickinson, M. E. et al. High-throughput discovery of novel developmental phenotypes. Nature 537, 508–514 (2016). This study uncovers mouse embryonic lethal (essential) genes and their relationship to human disease loci from large-scale phenotyping and analysis of hundreds of knockout mutations.
Chong, J. X. et al. The genetic basis of mendelian phenotypes: discoveries, challenges, and opportunities. Am. J. Hum. Genet. 97, 199–215 (2015).
Boyle, E. A., Li, Y. I. & Pritchard, J. K. An expanded view of complex traits: from polygenic to omnigenic. Cell 169, 1177–1186 (2017).
Bush, W. S., Oetjens, M. T. & Crawford, D. C. Unravelling the human genome-phenome relationship using phenome-wide association studies. Nat. Rev. Genet. 17, 129–145 (2016).
Schlager, G. & Dickie, M. M. Natural mutation rates in the house mouse. Estimates for five specific loci and dominant mutations. Mutat. Res. 11, 89–96 (1971).
Davisson, M. T., Bergstrom, D. E., Reinholdt, L. G. & Donahue, L. R. Discovery genetics - the history and future of spontaneous mutation research. Curr. Protoc. Mouse Biol. 2, 103–118 (2012).
Rogers, D. C. et al. Behavioral and functional analysis of mouse phenotype: SHIRPA, a proposed protocol for comprehensive phenotype assessment. Mamm. Genome 8, 711–713 (1997).
Rogers, D. C. et al. SHIRPA, a protocol for behavioral assessment: validation for longitudinal study of neurological dysfunction in mice. Neurosci. Lett. 306, 89–92 (2001).
Russell, L. B., Russell, W. L., Popp, R. A., Vaughan, C. & Jacobson, K. B. Radiation-induced mutations at mouse hemoglobin loci. Proc. Natl Acad. Sci. USA 73, 2843–2846 (1976).
Russell, W. L. et al. Specific-locus test shows ethylnitrosourea to be the most potent mutagen in the mouse. Proc. Natl Acad. Sci. USA 76, 5818–5819 (1979). This is a key publication describing the use of ENU to efficiently induce point mutations in mice and facilitate broad-based forward genetic screens.
Nolan, P. M. et al. Implementation of a large-scale ENU mutagenesis program: towards increasing the mouse mutant resource. Mamm. Genome 11, 500–506 (2000).
Arnold, C. N. et al. ENU-induced phenovariance in mice: inferences from 587 mutations. BMC Res. Notes 5, 577 (2012).
Oliver, P. L. & Davies, K. E. New insights into behaviour using mouse ENU mutagenesis. Hum. Mol. Genet. 21, R72–R81 (2012).
Boles, M. K. et al. A mouse chromosome 4 balancer ENU-mutagenesis screen isolates eleven lethal lines. BMC Genet. 10, 12 (2009).
Liu, X. et al. ENU mutagenesis screen to establish motor phenotypes in wild-type mice and modifiers of a pre-existing motor phenotype in tau mutant mice. J. Biomed. Biotechnol. 2011, 130947 (2011).
Tucci, V. et al. Reaching and grasping phenotypes in the mouse (Mus musculus): a characterization of inbred strains and mutant lines. Neuroscience 147, 573–582 (2007).
Zimprich, A. et al. Analysis of locomotor behavior in the German Mouse Clinic. J. Neurosci. Methods https://doi.org/10.1016/j.jneumeth.2017.05.005 (2017).
Wilson, L. et al. Random mutagenesis of proximal mouse chromosome 5 uncovers predominantly embryonic lethal mutations. Genome Res. 15, 1095–1105 (2005).
Flint, J. et al. A simple genetic basis for a complex psychological trait in laboratory mice. Science 269, 1432–1435 (1995).
Wada, Y. et al. ENU mutagenesis screening for dominant behavioral mutations based on normal control data obtained in home-cage activity, open-field, and passive avoidance tests. Exp. Anim. 59, 495–510 (2010).
Vitaterna, M. H. et al. Mutagenesis and mapping of a mouse gene, Clock, essential for circadian behavior. Science 264, 719–725 (1994).
Mandillo, S. et al. Reliability, robustness, and reproducibility in mouse behavioral phenotyping: a cross-laboratory study. Physiol. Genom. 34, 243–255 (2008). This study highlights the importance of cross-centre standardization and validation of phenotyping platforms.
Isaacs, A. M. et al. A mutation in Af4 is predicted to cause cerebellar ataxia and cataracts in the robotic mouse. J. Neurosci. 23, 1631–1637 (2003).
Clapcote, S. J. et al. Behavioral phenotypes of Disc1 missense mutations in mice. Neuron 54, 387–402 (2007).
Potter, P. K. et al. Novel gene function revealed by mouse mutagenesis screens for models of age-related disease. Nat. Commun. 7, 12444 (2016). Ageing and recurrent broad-based screening of mutants from large-scale mutagenesis programmes reveals novel gene functions underlying age-related disease.
Hardisty-Hughes, R. E., Parker, A. & Brown, S. D. A hearing and vestibular phenotyping pipeline to identify mouse mutants with hearing impairment. Nat. Protoc. 5, 177–190 (2010).
Esapa, C. T. et al. N-Ethyl-N-nitrosourea (ENU) induced mutations within the klotho gene lead to ectopic calcification and reduced lifespan in mouse models. PLOS One 10, e0122650 (2015).
Carpinelli, M. R. et al. Suppressor screen in Mpl−/− mice: c-Myb mutation causes supraphysiological production of platelets in the absence of thrombopoietin signaling. Proc. Natl Acad. Sci. USA 101, 6553–6558 (2004).
Aigner, B. et al. Diabetes models by screen for hyperglycemia in phenotype-driven ENU mouse mutagenesis projects. Am. J. Physiol. Endocrinol. Metab. 294, E232–E240 (2008).
Hough, T. A. et al. Novel phenotypes identified by plasma biochemical screening in the mouse. Mamm. Genome 13, 595–602 (2002).
Aigner, B. et al. Generation of N-ethyl-N-nitrosourea-induced mouse mutants with deviations in hematological parameters. Mamm. Genome 22, 495–505 (2011).
Hoebe, K. & Beutler, B. Forward genetic analysis of TLR-signaling pathways: an evaluation. Adv. Drug Deliv. Rev. 60, 824–829 (2008). This paper reviews how forward genetic mouse screens revealed the pathways that activate the innate immune system.
Miosge, L. A., Blasioli, J., Blery, M. & Goodnow, C. C. Analysis of an ethylnitrosourea-generated mouse mutation defines a cell intrinsic role of nuclear factor kappaB2 in regulating circulating B cell numbers. J. Exp. Med. 196, 1113–1119 (2002).
Nelms, K. A. & Goodnow, C. C. Genome-wide ENU mutagenesis to reveal immune regulators. Immunity 15, 409–418 (2001).
Adissu, H. A. et al. Histopathology reveals correlative and unique phenotypes in a high-throughput mouse phenotyping screen. Dis. Model. Mech. 7, 515–524 (2014).
Wong, M. D., Dorr, A. E., Walls, J. R., Lerch, J. P. & Henkelman, R. M. A novel 3D mouse embryo atlas based on micro-CT. Development 139, 3248–3256 (2012).
Wong, M. D., Maezawa, Y., Lerch, J. P. & Henkelman, R. M. Automated pipeline for anatomical phenotyping of mouse embryos using micro-CT. Development 141, 2533–2541 (2014).
Wong, M. D. et al. 4D atlas of the mouse embryo for precise morphological staging. Development 142, 3583–3591 (2015).
Weninger, W. J. et al. Phenotyping structural abnormalities in mouse embryos using high-resolution episcopic microscopy. Dis. Model. Mech. 7, 1143–1152 (2014).
Gailus-Durner, V. et al. Introducing the German Mouse Clinic: open access platform for standardized phenotyping. Nat. Methods 2, 403–404 (2005). This paper illuminates the important concept of the mouse clinic as a centre for mutant generation and broad-based phenotyping.
Brown, S. D., Chambon, P., de Angelis, M. H. & Eumorphia, C. EMPReSS: standardized phenotype screens for functional annotation of the mouse genome. Nat. Genet. 37, 1155 (2005).
Tucci, V. et al. Gene-environment interactions differentially affect mouse strain behavioral parameters. Mamm. Genome 17, 1113–1120 (2006).
Bradley, A. et al. The mammalian gene function resource: the International Knockout Mouse Consortium. Mamm. Genome 23, 580–586 (2012).
Skarnes, W. C. et al. A conditional knockout resource for the genome-wide study of mouse gene function. Nature 474, 337–342 (2011). References 56 and 57 describe the IKMC mouse mutant resource, which has been key to the development of high-throughput mouse phenomics.
Blair, D. R. et al. A nondegenerate code of deleterious variants in Mendelian loci contributes to complex disease risk. Cell 155, 70–80 (2013). By data mining medical records of over 100 million patients, the authors revealed a correlation between rare genetic diseases and complex disease, demonstrating that highly penetrant phenotypic alleles can help understand the genetic aetiology of common disorders.
Sungur, A. O., Schwarting, R. K. & Wohr, M. Early communication deficits in the Shank1 knockout mouse model for autism spectrum disorder: developmental aspects and effects of social context. Autism Res. 9, 696–709 (2016).
Sungur, A. O., Schwarting, R. K. W. & Wohr, M. Behavioral phenotypes and neurobiological mechanisms in the Shank1 mouse model for autism spectrum disorder: A translational perspective. Behav. Brain Res. https://doi.org/10.1016/j.bbr.2017.09.038 (2017).
Schmeisser, M. J. et al. Autistic-like behaviours and hyperactivity in mice lacking ProSAP1/Shank2. Nature 486, 256–260 (2012).
Katayama, Y. et al. CHD8 haploinsufficiency results in autistic-like phenotypes in mice. Nature 537, 675–679 (2016).
Platt, R. J. et al. Chd8 mutation leads to autistic-like behaviors and impaired striatal circuits. Cell Rep. 19, 335–350 (2017).
Meehan, T. F. et al. Disease model discovery from 3,328 gene knockouts by The International Mouse Phenotyping Consortium. Nat. Genet. 49, 1231–1238 (2017). This study provides extensive novel insights into gene function along with numerous new disease models from the work of the IMPC.
Raj, A., Rifkin, S. A., Andersen, E. & van Oudenaarden, A. Variability in gene expression underlies incomplete penetrance. Nature 463, 913–918 (2010).
Karp, N. A. et al. Prevalence of sexual dimorphism in mammalian phenotypic traits. Nat. Commun. 8, 15475 (2017). In this large study involving over 50,000 mice, the authors demonstrate that differences exist between male and female mice in the majority of phenotypic screens employed by the IMPC.
Bowl, M. R. et al. A large scale hearing loss screen reveals an extensive unexplored genetic landscape for auditory dysfunction. Nat. Commun. 8, 886 (2017).
Rozman, J. et al. Identification of genetic elements in metabolism by high-throughput mouse phenotyping. Nat. Commun. 9, 288 (2018).
Peirce, J. L., Lu, L., Gu, J., Silver, L. M. & Williams, R. W. A new set of BXD recombinant inbred lines from advanced intercross populations in mice. BMC Genet. 5, 7 (2004).
Churchill, G. A. et al. The Collaborative Cross, a community resource for the genetic analysis of complex traits. Nat. Genet. 36, 1133–1137 (2004). This paper describes plans to create hundreds of independently bred, recombinant inbred mouse lines from eight inbred parental strains to study polygenic networks and interactions among genes that complement knockout mouse studies.
Paigen, K. & Eppig, J. T. A mouse phenome project. Mamm. Genome 11, 715–717 (2000).
Bennett, B. J. et al. A high-resolution association mapping panel for the dissection of complex traits in mice. Genome Res. 20, 281–290 (2010).
Patterson, M. et al. Frequency of mononuclear diploid cardiomyocytes underlies natural variation in heart regeneration. Nat. Genet. 49, 1346–1353 (2017).
Shusterman, A. et al. Genotype is an important determinant factor of host susceptibility to periodontitis in the collaborative cross and inbred mouse populations. BMC Genet. 14, 68 (2013).
Mao, J. H. et al. Identification of genetic factors that modify motor performance and body weight using collaborative cross mice. Sci. Rep. 5, 16247 (2015).
Solberg, L. C. et al. A protocol for high-throughput phenotyping, suitable for quantitative trait analysis in mice. Mamm. Genome 17, 129–146 (2006).
Valdar, W. et al. Genome-wide genetic association of complex traits in heterogeneous stock mice. Nat. Genet. 38, 879–887 (2006).
Svenson, K. L. et al. High-resolution genetic mapping using the mouse diversity outbred population. Genetics 190, 437–447 (2012).
Churchill, G. A., Gatti, D. M., Munger, S. C. & Svenson, K. L. The diversity outbred mouse population. Mamm. Genome 23, 713–718 (2012). A variation of the CC line that allows population genetic studies using a heterogeneous stock where each mouse is genetically unique and the extent of genetic variability is similar to that observed in humans.
Pallares, L. F. et al. Mapping of craniofacial traits in outbred mice identifies major developmental genes involved in shape determination. PLOS Genet. 11, e1005607 (2015).
Nicod, J. et al. Genome-wide association of multiple complex traits in outbred mice by ultra-low-coverage sequencing. Nat. Genet. 48, 912–918 (2016). This study reports comprehensive phenotyping pipelines applied to the genetic analysis of an outbred mouse population, revealing numerous complex traits mapped at gene-level resolution.
Doetschman, T. Influence of genetic background on genetically engineered mouse phenotypes. Methods Mol. Biol. 530, 423–433 (2009).
Broman, K. W. The genomes of recombinant inbred lines. Genetics 169, 1133–1146 (2005).
Sittig, L. J. et al. Genetic background limits generalizability of genotype-phenotype relationships. Neuron 91, 1253–1259 (2016).
Moon, C. et al. Vertically transmitted faecal IgA levels determine extra-chromosomal phenotypic variation. Nature 521, 90–93 (2015).
Nguyen, T. L., Vieira-Silva, S., Liston, A. & Raes, J. How informative is the mouse for human gut microbiota research? Dis. Model. Mech. 8, 1–16 (2015).
Overton, J. M. Phenotyping small animals as models for the human metabolic syndrome: thermoneutrality matters. Int. J. Obes. 34, (Suppl. 2) S53–S58 (2010).
Robinson, L. & Riedel, G. Comparison of automated home-cage monitoring systems: emphasis on feeding behaviour, activity and spatial learning following pharmacological interventions. J. Neurosci. Methods 234, 13–25 (2014).
Spruijt, B. M., Peters, S. M., de Heer, R. C., Pothuizen, H. H. & van der Harst, J. E. Reproducibility and relevance of future behavioral sciences should benefit from a cross fertilization of past recommendations and today’s technology: “Back to the future”. J. Neurosci. Methods 234, 2–12 (2014).
Bains, R. S. et al. Assessing mouse behaviour throughout the light/dark cycle using automated in-cage analysis tools. J. Neurosci. Methods. https://doi.org/10.1016/j.jneumeth.2017.04.014 (2017).
Hong, W. et al. Automated measurement of mouse social behaviors using depth sensing, video tracking, and machine learning. Proc. Natl Acad. Sci. USA 112, E5351–E5360 (2015).
Zarringhalam, K. et al. An open system for automatic home-cage behavioral analysis and its application to male and female mouse models of Huntington’s disease. Behav. Brain Res. 229, 216–225 (2012).
Steele, A. D., Jackson, W. S., King, O. D. & Lindquist, S. The power of automated high-resolution behavior analysis revealed by its application to mouse models of Huntington’s and prion diseases. Proc. Natl Acad. Sci. USA 104, 1983–1988 (2007).
Bains, R. S. et al. Analysis of individual mouse activity in group housed animals of different inbred strains using a novel automated home cage analysis system. Front. Behav. Neurosci. 10, 106 (2016).
Valletta, J. J., Torney, C., Kings, M., Thornton, A. & Madden, J. Applications of machine learning in animal behaviour studies. Animal Behav. 124, 203–220 (2017).
Atamni, H. J., Mott, R., Soller, M. & Iraqi, F. A. High-fat-diet induced development of increased fasting glucose levels and impaired response to intraperitoneal glucose challenge in the collaborative cross mouse genetic reference population. BMC Genet. 17, 10 (2016).
Myint, A. et al. Large-scale phenotyping of noise-induced hearing loss in 100 strains of mice. Hear Res. 332, 113–120 (2016).
Ferris, M. T. et al. Modeling host genetic regulation of influenza pathogenesis in the collaborative cross. PLOS Pathog. 9, e1003196 (2013).
Horsch, M. et al. Cox4i2, Ifit2, and Prdm11 mutant mice: effective selection of genes predisposing to an altered airway inflammatory response from a large compendium of mutant mouse lines. PLOS One 10, e0134503 (2015).
Sundberg, J. P. et al. The mouse as a model for understanding chronic diseases of aging: the histopathologic basis of aging in inbred mice. Pathobiol Aging Age Relat. Dis. 1, https://doi.org/10.3402/pba.v1i0.7179 (2011).
Sundberg, J. P. et al. Approaches to investigating complex genetic traits in a large-scale inbred mouse aging study. Vet. Pathol. 53, 456–467 (2016).
Karp, N. A. et al. Applying the ARRIVE guidelines to an in vivo database. PLOS Biol. 13, e1002151 (2015).
Karp, N. A. et al. Impact of temporal variation on design and analysis of mouse knockout phenotyping studies. PLOS One 9, e111239 (2014).
Van Valen, D. A. et al. Deep learning automates the quantitative analysis of individual cells in live-cell imaging experiments. PLOS Comput. Biol. 12, e1005177 (2016).
Wilkinson, M. D. et al. The FAIR guiding principles for scientific data management and stewardship. Sci. Data 3, 160018 (2016).
Roadmap Epigenomics Consortium et al. Integrative analysis of 111 reference human epigenomes. Nature 518, 317–330 (2015). This resource describes the collection, archiving and analysis of epigenomic data generated as part of the NIH Roadmap Epigenomics Consortium.
Kawaji, H., Kasukawa, T., Forrest, A., Carninci, P. & Hayashizaki, Y. The FANTOM5 collection, a data series underpinning mammalian transcriptome atlases in diverse cell types. Sci. Data 4, 170113 (2017). This paper reviews the wide impact the fifth iteration of the RIKEN-led FANTOM consortium has had on understanding cell function by the generation of a comprehensive cellular transcription atlas.
Sudlow, C. et al. UK biobank: an open access resource for identifying the causes of a wide range of complex diseases of middle and old age. PLOS Med. 12, e1001779 (2015). This is a large-scale resource that integrates extensive phenotypic and genotypic data from >500,000 participants to support investigations into the genetic and non-genetic determinants of the diseases of middle and old age.
Smith, C. L. & Eppig, J. T. The mammalian phenotype ontology as a unifying standard for experimental and high-throughput phenotyping data. Mamm. Genome 23, 653–668 (2012).
Hayamizu, T. F. et al. EMAP/EMAPA ontology of mouse developmental anatomy: 2013 update. J. Biomed. Semant. 4, 15 (2013).
Gkoutos, G. V., Schofield, P. N. & Hoehndorf, R. The anatomy of phenotype ontologies: principles, properties and applications. Brief Bioinform. https://doi.org/10.1093/bib/bbx035 (2017).
Consortium, G. T. et al. Genetic effects on gene expression across human tissues. Nature 550, 204–213 (2017).
Szklarczyk, D. et al. The STRING database in 2017: quality-controlled protein-protein association networks, made broadly accessible. Nucleic Acids Res. 45, D362–D368 (2017).
Rath, A. et al. Representation of rare diseases in health information systems: the Orphanet approach to serve a wide range of end users. Hum. Mutat. 33, 803–808 (2012).
Zhu, X. & Stephens, M. A large-scale genome-wide enrichment analysis identifies new trait-associated genes, pathways and tissues across 31 human phenotypes. bioRxiv. https://doi.org/10.1101/160770 (2017).
Rukat, T., Holmes, C. C., Titsias, M. K. & Yau, C. Bayesian boolean matrix factorisation. Proc. Machine Learn. Res. 70, 2969–2978 (2017).
Wiltschko, A. B. et al. Mapping sub-second structure in mouse behavior. Neuron 88, 1121–1135 (2015).
Cortes, A. et al. Bayesian analysis of genetic association across tree-structured routine healthcare data in the UK Biobank. Nat. Genet. 49, 1311–1318 (2017).
Ng, S. B. et al. Exome sequencing identifies the cause of a mendelian disorder. Nat. Genet. 42, 30–35 (2010).
Rabbani, B., Mahdieh, N., Hosomichi, K., Nakaoka, H. & Inoue, I. Next-generation sequencing: impact of exome sequencing in characterizing Mendelian disorders. J. Hum. Genet. 57, 621–632 (2012).
Ramoni, R. B. et al. The undiagnosed diseases network: accelerating discovery about health and disease. Am. J. Hum. Genet. 100, 185–192 (2017).
Collins, F. S. & Varmus, H. A new initiative on precision medicine. N. Engl. J. Med. 372, 793–795 (2015). This is an announcement of how the NIH will refocus efforts in the precision medicine era, including the need for more reliable models for preclinical testing.
Yang, Y. et al. Clinical whole-exome sequencing for the diagnosis of mendelian disorders. N. Engl. J. Med. 369, 1502–1511 (2013).
Yang, Y. et al. Molecular findings among patients referred for clinical whole-exome sequencing. JAMA 312, 1870–1879 (2014).
Lee, H. et al. Clinical exome sequencing for genetic identification of rare Mendelian disorders. JAMA 312, 1880–1887 (2014).
Koscielny, G. et al. The International Mouse Phenotyping Consortium Web Portal, a unified point of access for knockout mice and related phenotyping data. Nucleic Acids Res. 42, D802–D809 (2014).
Wang, J. et al. MARRVEL: Integration of human and model organism genetic resources to facilitate functional annotation of the human genome. Am. J. Hum. Genet. 100, 843–853 (2017).
Mungall, C. J. et al. The monarch initiative: an integrative data and analytic platform connecting phenotypes to genotypes across species. Nucleic Acids Res. 45, D712–D722 (2017).
Smedley, D. & Robinson, P. N. Phenotype-driven strategies for exome prioritization of human Mendelian disease genes. Genome Med. 7, 81 (2015).
Robinson, P. N. et al. Improved exome prioritization of disease genes through cross-species phenotype comparison. Genome Res. 24, 340–348 (2014). This is an important algorithmic approach that uses genotype-to-phenotype data from model organism studies to assess the potential impact of exome variants identified in sequencing from patients with a rare disease.
Smedley, D. et al. A whole-genome analysis framework for effective identification of pathogenic regulatory variants in Mendelian disease. Am. J. Hum. Genet. 99, 595–606 (2016).
Singleton, M. V. et al. Phevor combines multiple biomedical ontologies for accurate identification of disease-causing alleles in single individuals and small nuclear families. Am. J. Hum. Genet. 94, 599–610 (2014).
Gall, T. et al. Defining disease, diagnosis, and translational medicine within a homeostatic perturbation paradigm: the national institutes of health undiagnosed diseases program experience. Front. Med. 4, 62 (2017).
Kohler, S. et al. The human phenotype ontology in 2017. Nucleic Acids Res. 45, D865–D876 (2017).
Mungall, C. J. et al. Integrating phenotype ontologies across multiple species. Genome Biol. 11, R2 (2010).
Acknowledgements
The authors are grateful to J. McMurry for help with Fig. 6, which is a composition of several images previously licensed under CC0 in Pixabay and some under CC-BY 4.0 in https://github.com/jmcmurry/open-illustrations. The authors also thank their colleagues in the International Mouse Phenotyping Consortium, who have contributed in no small measure to the consideration and future of mouse phenomics. The authors thank many colleagues who have participated with them in other consortia involving mouse phenomics, including European Union Mouse Genetics Research for Public Health and Industrial Applications (EUMORPHIA), European Mouse Disease Clinic (EUMODIC) and European Conditional Mouse Mutagenesis Program (EUCOMM). The views expressed in this article are based on the many discussions and insights that have emerged within these consortia over many years. Lastly, the authors are grateful to the Medical Research Council, UK (S.D.M.B., C.C.H., A-M.M. and S.W.), the National Institutes of Health Grant UM1-HG006370 (A-M.M., T.F.M. and D.S.), and the Engineering and Physical Sciences Research Council, UK (C.C.H.) for funding support.
Reviewer information
Nature Reviews thanks Jonathan Flint, K. C. Kent Lloyd and the other anonymous reviewer(s), for their contribution to the peer review of this work.
Author information
Authors and Affiliations
Contributions
All authors contributed to all aspects of this manuscript, including researching data, discussing content, writing, and reviewing and editing the manuscript before submission.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Related links
International Mouse Phenotyping Consortium (IMPC): http://www.mousephenotype.org
International Mouse Phenotyping Resource of Standardised Screens (IMPReSS): http://www.mousephenotype.org
Online Mendelian Inheritance in Man (OMIM): http://www.omim.org
Orphanet: http://www.orpha.net
Hybrid Mouse Diversity Panel (HMDP): https://systems.genetics.ucla.edu/
Edinburgh Mouse Atlas Project (EMAP): http://www.emouseatlas.org/
Phenotype And Trait Ontology (PATO): https://bioportal.bioontology.org/ontologies/PATO
Genotype–Tissue Expression (GTEx) Project: https://www.gtexportal.org/home/
Search Tool for the Retrieval of Interacting Genes/Proteins (STRING): https://string-db.org/
Monarch Initiative: https://monarchinitiative.org/
100,000 Genomes Project: https://www.genomicsengland.co.uk/
NIH Undiagnosed Diseases Network (UDN): https://undiagnosed.hms.harvard.edu/
Mouse Genome Informatics: http://www.informatics.jax.org
Glossary
- Variable expressivity
-
Differing phenotypic features among individuals with the same genotype.
- Phenotypic expansion
-
The expanding array of phenotypes that may be associated with mutations in a specific gene.
- Genome-wide association studies
-
(GWAS). Genome-wide analyses of single nucleotide polymorphisms (SNPs) in human cohorts to test for associations between SNPs and traits.
- Phenome-wide association studies
-
(PheWAS). Testing genetic variants for an association with multiple phenotypes or traits (the phenome) in human cohorts.
- Pre-pulse inhibition
-
(PPI). Used to assess sensorimotor gating. In the PPI test, sensorimotor gating is assessed by measuring the innate reduction of the startle reflex induced by a weak prestimulus (prepulse) before a subsequent strong startle stimulus (pulse). Deficits in PPI responses are noted in patients suffering from a range of illnesses, including schizophrenia.
- Optokinetic drum
-
Assesses the threshold of visual acuity by placing a mouse in the centre of a rotating drum and measuring reflexive head turning in response to the rotation of stripes, which subsequently decrease in width and distance of separation.
- Auditory brainstem response
-
Measures the electrical response in the auditory nerve and brainstem to either a defined frequency or a longer, complex auditory stimulus. This allows frequency-specific auditory thresholds to be determined.
- Gene trapping
-
A random insertional mutation into an intron or exon of a gene that disrupts expression of the trapped gene.
- Gene targeting
-
Targeting by homologous recombination into embryonic stem cells to introduce mutations ranging from single base pair substitutions to large deletions.
- Endophenotypes
-
A heritable and measurable component of a phenotype, which is intermediate between gene and disease.
- Coisogenic
-
Isogenic strains differing only at a single locus. Thus, all International Mouse Phenotyping Consortium (IMPC) lines are coisogenic on the C57BL/6 N background.
- Optical projection tomography
-
(OPT). An optical computed tomography technique that is used to acquire 3D images of early embryo morphology in the mouse.
- Micro-computed tomography
-
(µCT). High-resolution X-ray computed tomography to acquire 3D images of embryo morphology in the mouse, usually during later stages of development.
- High-resolution episcopic microscopy
-
(HREM). A method for the determination of the 3D structure of embryos using recurrent block surface (episcopic) imaging of sections from histological samples.
- Subviable lines
-
Mouse mutant lines for which some individual mice show embryonic lethality, whereas others of identical genotype survive.
- Paralogue
-
Paralogues are pairs of genes that derive from a common ancestral gene and may undertake similar functions.
- Recombinant inbred
-
(RI). Mouse lines that are derived by the intercrossing and subsequent inbreeding of two distinct inbred lines. Each line carries a differing patchwork of chromosome segments from the two parental lines, allowing researchers to relate phenotypic differences between the parental inbred strains to the underlying genetic loci involved.
- Collaborative Cross
-
(CC). Mouse lines that are a multi-parental recombinant inbred panel derived from crosses between eight inbred lines (including three wild-derived inbred strains), capturing a greater genetic diversity more evenly spread across the genome.
- Quantitative trait locus
-
(QTL). A locus that contributes some proportion of the total phenotypic variance of the quantitative trait. Many quantitative traits are determined by multiple genes (or QTLs), each of which may have small or large effects on the phenotype.
- Heterogeneous Stock
-
(HS). A type of mouse population that enables fine-resolution mapping of traits and is created by the intercrossing of inbred or recombinant inbred lines followed by mating schemes that minimize inbreeding.
- Diversity Outbred
-
(DO). A mouse population that is a Heterogeneous Stock that was derived by random mating of 144 partially inbred Collaborative Cross lines, providing single-gene mapping resolution.
- Ontology
-
Phenotype ontologies encompass the naming, description and interrelationship of phenotypes.
- Orphan drugs
-
Drugs that are developed to treat a rare medical condition, an orphan disease.
Rights and permissions
About this article
Cite this article
Brown, S.D.M., Holmes, C.C., Mallon, AM. et al. High-throughput mouse phenomics for characterizing mammalian gene function. Nat Rev Genet 19, 357–370 (2018). https://doi.org/10.1038/s41576-018-0005-2
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41576-018-0005-2
This article is cited by
-
How is Big Data reshaping preclinical aging research?
Lab Animal (2023)
-
EmbryoNet: using deep learning to link embryonic phenotypes to signaling pathways
Nature Methods (2023)
-
Pleiotropy data resource as a primer for investigating co-morbidities/multi-morbidities and their role in disease
Mammalian Genome (2022)
-
Progress towards completing the mutant mouse null resource
Mammalian Genome (2022)
-
A Simple Evolutionary Model of Genetic Robustness After Gene Duplication
Journal of Molecular Evolution (2022)