Abstract
Nonalcoholic fatty liver disease (NAFLD) is one of the most common liver diseases worldwide and is often associated with aspects of metabolic syndrome. Despite its prevalence and the importance of early diagnosis, there is a lack of robustly validated biomarkers for diagnosis, prognosis and monitoring of disease progression in response to a given treatment. In this Review, we provide an overview of the contribution of metabolomics and lipidomics in clinical studies to identify biomarkers associated with NAFLD and nonalcoholic steatohepatitis (NASH). In addition, we highlight the key metabolic pathways in NAFLD and NASH that have been identified by metabolomics and lipidomics approaches and could potentially be used as biomarkers for non-invasive diagnostic tests. Overall, the studies demonstrated alterations in amino acid metabolism and several aspects of lipid metabolism including circulating fatty acids, triglycerides, phospholipids and bile acids. Although we report several studies that identified potential biomarkers, few have been validated.
Key points
-
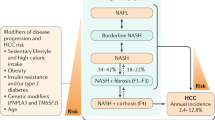
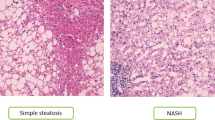
Nonalcoholic fatty liver disease (NAFLD) affects 25% of the adult world population; in about 20% of patients, it can progress to nonalcoholic steatohepatitis (NASH), which can lead to cirrhosis.
-
There is an urgent need for development of clinically relevant biomarkers and non-invasive diagnostic tests for NAFLD.
-
Metabolomics and lipidomics approaches have provided insightful evidence of altered metabolic pathways in NAFLD and NASH.
-
There is an association between circulating amino acids and steatohepatitis, and impairment in amino acid metabolism in NAFLD is strongly correlated with insulin resistance, particularly in the muscle.
-
An increase in oxidative stress results in a reduction in hepatic glutathione levels and is associated with liver damage and the progression of NAFLD to NASH.
-
NASH is strongly associated with alterations in circulating fatty acids and intact lipids, which is partially due to alterations in de novo liver lipogenesis, lipolysis rate and VLDL metabolism.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Younossi, Z. M. et al. Global epidemiology of nonalcoholic fatty liver disease-meta-analytic assessment of prevalence, incidence, and outcomes. Hepatology 64, 73–84 (2016).
Younossi, Z. et al. Global burden of NAFLD and NASH: trends, predictions, risk factors and prevention. Nat. Rev. Gastroenterol. Hepatol. 15, 11–20 (2018).
FDA-NIH Biomarker Working Group. BEST (Biomarkers, EndpointS, and other Tools) Resource [Internet] (FDA, 2016).
Silverman, J. F. et al. Liver pathology in morbidly obese patients with and without diabetes. Am. J. Gastroenterol. 85, 1349–1355 (1990).
Loguercio, C. et al. Non-alcoholic fatty liver disease in an area of southern Italy: main clinical, histological, and pathophysiological aspects. J. Hepatol. 35, 568–574 (2001).
Neuschwander-Tetri, B. A. & Bacon, B. R. Nonalcoholic steatohepatitis. Med. Clin. North Am. 80, 1147–1166 (1996).
Muriel, P. Role of free radicals in liver diseases. Hepatol. Int. 3, 526–536 (2009).
Rinella, M. E., Tacke, F., Sanyal, A. J., Anstee, Q. M. & Participants of the AASLD/EASL Workshop. Report on the AASLD/EASL joint workshop on clinical trial endpoints in NAFLD. J. Hepatol. 71, 823–833 (2019).
Ratziu, V. A critical review of endpoints for non-cirrhotic NASH therapeutic trials. J. Hepatol. 68, 353–361 (2018).
European Association for the Study of the Liver (EASL), European Association for the Study of Diabetes (EASD) & European Association for the Study of Obesity (EASO). EASL-EASD-EASO Clinical Practice Guidelines for the management of non-alcoholic fatty liver disease. J. Hepatol. 64, 1388–1402 (2016).
Hoofnagle, J. H. et al. Vitamin E and changes in serum alanine aminotransferase levels in patients with non-alcoholic steatohepatitis. Aliment. Pharmacol. Ther. 38, 134–143 (2013).
Vilar-Gomez, E. et al. Development and validation of a noninvasive prediction model for nonalcoholic steatohepatitis resolution after lifestyle intervention. Hepatology 63, 1875–1887 (2016).
Shah, A. G. et al. Comparison of noninvasive markers of fibrosis in patients with nonalcoholic fatty liver disease. Clin. Gastroenterol. Hepatol. 7, 1104–1112 (2009).
Quehenberger, O. et al. Lipidomics reveals a remarkable diversity of lipids in human plasma. J. Lipid Res. 51, 3299–3305 (2010).
Hyotylainen, T. & Oresic, M. Systems biology strategies to study lipidomes in health and disease. Prog. Lipid Res. 55, 43–60 (2014).
Cajka, T. & Fiehn, O. Toward merging untargeted and targeted methods in mass spectrometry-based metabolomics and lipidomics. Anal. Chem. 88, 524–545 (2016).
Mardinoglu, A. et al. Genome-scale metabolic modelling of hepatocytes reveals serine deficiency in patients with non-alcoholic fatty liver disease. Nat. Commun. 5, 3083 (2014).
Mardinoglu, A. et al. Personal model-assisted identification of NAD(+) and glutathione metabolism as intervention target in NAFLD. Mol. Syst. Biol. 13, 916 (2017).
Hyotylainen, T. et al. Genome-scale study reveals reduced metabolic adaptability in patients with non-alcoholic fatty liver disease. Nat. Commun. 7, 8994 (2016).
Nielsen, J. Systems biology of metabolism: a driver for developing personalized and precision medicine. Cell Metab. 25, 572–579 (2017).
Kalhan, S. C. et al. Plasma metabolomic profile in nonalcoholic fatty liver disease. Metabolism 60, 404–413 (2011).
Lake, A. D. et al. Branched chain amino acid metabolism profiles in progressive human nonalcoholic fatty liver disease. Amino Acids 47, 603–615 (2015).
Gaggini, M. et al. Altered amino acid concentrations in NAFLD: impact of obesity and insulin resistance. Hepatology 67, 145–158 (2018).
Kakazu, E. et al. Branched chain amino acids are associated with the heterogeneity of the area of lipid droplets in hepatocytes of patients with non-alcoholic fatty liver disease. Hepatol. Res. 49, 860–871 (2019).
Barr, J. et al. Obesity-dependent metabolic signatures associated with nonalcoholic fatty liver disease progression. J. Proteome Res. 11, 2521–2532 (2012).
Biolo, G., Gastaldelli, A., Zhang, X. J. & Wolfe, R. R. Protein synthesis and breakdown in skin and muscle: a leg model of amino acid kinetics. Am. J. Physiol. 267, E467–E474 (1994).
Lynch, C. J. & Adams, S. H. Branched-chain amino acids in metabolic signalling and insulin resistance. Nat. Rev. Endocrinol. 10, 723–736 (2014).
Fiehn, O. et al. Plasma metabolomic profiles reflective of glucose homeostasis in non-diabetic and type 2 diabetic obese African-American women. PLoS ONE 5, e15234 (2010).
Newgard, C. B. et al. A branched-chain amino acid-related metabolic signature that differentiates obese and lean humans and contributes to insulin resistance. Cell Metab. 9, 311–326 (2009).
Lee, Y. H. et al. Sarcopenia is associated with significant liver fibrosis independently of obesity and insulin resistance in nonalcoholic fatty liver disease: nationwide surveys (KNHANES 2008–2011). Hepatology 63, 776–786 (2016).
Adeva, M. M., Calvino, J., Souto, G. & Donapetry, C. Insulin resistance and the metabolism of branched-chain amino acids in humans. Amino Acids 43, 171–181 (2012).
Wang, T. J. et al. Metabolite profiles and the risk of developing diabetes. Nat. Med. 17, 448–453 (2011).
Luzi, L., Castellino, P. & DeFronzo, R. A. Insulin and hyperaminoacidemia regulate by a different mechanism leucine turnover and oxidation in obesity. Am. J. Physiol. 270, E273–E281 (1996).
Qi, S. et al. Metabonomics screening of serum identifies pyroglutamate as a diagnostic biomarker for nonalcoholic steatohepatitis. Clin. Chim. Acta 473, 89–95 (2017).
Dong, S. et al. Urinary metabolomics analysis identifies key biomarkers of different stages of nonalcoholic fatty liver disease. World J. Gastroenterol. 23, 2771–2784 (2017).
Kitajima, Y. et al. Supplementation with branched-chain amino acids ameliorates hypoalbuminemia, prevents sarcopenia, and reduces fat accumulation in the skeletal muscles of patients with liver cirrhosis. J. Gastroenterol. 53, 427–437 (2018).
Kantartzis, K. et al. High cardiorespiratory fitness is an independent predictor of the reduction in liver fat during a lifestyle intervention in non-alcoholic fatty liver disease. Gut 58, 1281–1288 (2009).
Lehmann, R. et al. Circulating lysophosphatidylcholines are markers of a metabolically benign nonalcoholic fatty liver. Diabetes Care 36, 2331–2338 (2013).
Morgan, M. Y., Marshall, A. W., Milsom, J. P. & Sherlock, S. Plasma amino-acid patterns in liver disease. Gut 23, 362–370 (1982).
Morgan, M. Y., Milsom, J. P. & Sherlock, S. Plasma ratio of valine, leucine and isoleucine to phenylalanine and tyrosine in liver disease. Gut 19, 1068–1073 (1978).
Fischer, J. E. et al. The effect of normalization of plasma amino acids on hepatic encephalopathy in man. Surgery 80, 77–91 (1976).
Michitaka, K. et al. Amino acid imbalance in patients with chronic liver diseases. Hepatol. Res. 40, 393–398 (2010).
Ishikawa, T. et al. Branched-chain amino acids to tyrosine ratio (BTR) predicts intrahepatic distant recurrence and survival for early hepatocellular carcinoma. Hepatogastroenterology 60, 2055–2059 (2013).
Matthews, D. E. An overview of phenylalanine and tyrosine kinetics in humans. J. Nutr. 137, 1549S–1555S; discussion 1573S–1575S (2007).
Haufe, S. et al. Branched-chain and aromatic amino acids, insulin resistance and liver specific ectopic fat storage in overweight to obese subjects. Nutr. Metab. Cardiovasc. Dis. 26, 637–642 (2016).
Kawanaka, M. et al. Tyrosine levels are associated with insulin resistance in patients with nonalcoholic fatty liver disease. Hepat. Med. 7, 29–35 (2015).
Kahl, S. et al. Amino acid and fatty acid levels affect hepatic phosphorus metabolite content in metabolically healthy humans. J. Clin. Endocrinol. Metab. 103, 460–468 (2018).
Gastaldelli, A. et al. Relationship between hepatic/visceral fat and hepatic insulin resistance in nondiabetic and type 2 diabetic subjects. Gastroenterology 133, 496–506 (2007).
McCullough, A., Previs, S. & Kasumov, T. Stable isotope-based flux studies in nonalcoholic fatty liver disease. Pharmacol. Ther. 181, 22–33 (2018).
Zhu, L. et al. Upregulation of non-canonical Wnt ligands and oxidative glucose metabolism in NASH induced by methionine-choline deficient diet. Trends Cell Mol. Biol. 13, 47–56 (2018).
Zhou, Y. et al. Noninvasive detection of nonalcoholic steatohepatitis using clinical markers and circulating levels of lipids and metabolites. Clin. Gastroenterol. Hepatol. 14, 1463–1472.e6 (2016).
Dasarathy, S. et al. Glycine and urea kinetics in nonalcoholic steatohepatitis in human: effect of intralipid infusion. Am. J. Physiol. Gastrointest. Liver Physiol 297, G567–G575 (2009).
Kalhan, S. C. et al. Methionine and protein metabolism in non-alcoholic steatohepatitis: evidence for lower rate of transmethylation of methionine. Clin. Sci. 121, 179–189 (2011).
Mato, J. M., Alonso, C., Noureddin, M. & Lu, S. C. Biomarkers and subtypes of deranged lipid metabolism in non-alcoholic fatty liver disease. World J. Gastroenterol. 25, 3009–3020 (2019).
Obeid, R. & Herrmann, W. Homocysteine and lipids: S-adenosyl methionine as a key intermediate. FEBS Lett. 583, 1215–1225 (2009).
Pastore, A. et al. Plasma levels of homocysteine and cysteine increased in pediatric NAFLD and strongly correlated with severity of liver damage. Int. J. Mol. Sci. 15, 21202–21214 (2014).
Lu, S. C. & Mato, J. M. S-adenosylmethionine in liver health, injury, and cancer. Physiol. Rev. 92, 1515–1542 (2012).
Alonso, C. et al. Metabolomic identification of subtypes of nonalcoholic steatohepatitis. Gastroenterology 152, 1449–1461.e7 (2017).
Sookoian, S. et al. Nonalcoholic steatohepatitis is associated with a state of betaine-insufficiency. Liver Int. 37, 611–619 (2017).
Zhang, W. et al. Betaine protects against high-fat-diet-induced liver injury by inhibition of high-mobility group Box 1 and Toll-like receptor 4 expression in rats. Dig. Dis. Sci. 58, 3198–3206 (2013).
Du, K. et al. Increased glutaminolysis marks active scarring in nonalcoholic steatohepatitis progression. Cell Mol. Gastroenterol. Hepatol. 10, 1–21 (2020).
Soga, T. et al. Serum metabolomics reveals γ-glutamyl dipeptides as biomarkers for discrimination among different forms of liver disease. J. Hepatol. 55, 896–905 (2011).
Koch, M. et al. Serum metabolomic profiling highlights pathways associated with liver fat content in a general population sample. Eur. J. Clin. Nutr. 71, 995–1001 (2017).
Cheng, J., Joyce, A., Yates, K., Aouizerat, B. & Sanyal, A. J. Metabolomic profiling to identify predictors of response to vitamin E for non-alcoholic steatohepatitis (NASH). PLoS ONE 7, e44106 (2012).
Leonetti, S., Herzog, R. I., Caprio, S., Santoro, N. & Tricò, D. Glutamate–serine–glycine index: a novel potential biomarker in pediatric non-alcoholic fatty liver disease. Children 7, 270 (2020).
Fabbrini, E. et al. Alterations in adipose tissue and hepatic lipid kinetics in obese men and women with nonalcoholic fatty liver disease. Gastroenterology 134, 424–431 (2008).
Cohen, J. C., Horton, J. D. & Hobbs, H. H. Human fatty liver disease: old questions and new insights. Science 332, 1519–1523 (2011).
Westerbacka, J. et al. Splanchnic balance of free fatty acids, endocannabinoids, and lipids in subjects with nonalcoholic fatty liver disease. Gastroenterology 139, 1961–1971.e1 (2010).
Donnelly, K. L. et al. Sources of fatty acids stored in liver and secreted via lipoproteins in patients with nonalcoholic fatty liver disease. J. Clin. Invest. 115, 1343–1351 (2005).
Bugianesi, E. et al. Insulin resistance in non-diabetic patients with non-alcoholic fatty liver disease: sites and mechanisms. Diabetologia 48, 634–642 (2005).
Lomonaco, R. et al. Effect of adipose tissue insulin resistance on metabolic parameters and liver histology in obese patients with nonalcoholic fatty liver disease. Hepatology 55, 1389–1397 (2012).
Rosso, C. et al. Crosstalk between adipose tissue insulin resistance and liver macrophages in non-alcoholic fatty liver disease. J. Hepatol. 71, 1012–1021 (2019).
Bell, L. N. et al. Relationship between adipose tissue insulin resistance and liver histology in nonalcoholic steatohepatitis: a pioglitazone versus vitamin E versus placebo for the treatment of nondiabetic patients with nonalcoholic steatohepatitis trial follow-up study. Hepatology 56, 1311–1318 (2012).
Gastaldelli, A. et al. Importance of changes in adipose tissue insulin resistance to histological response during thiazolidinedione treatment of patients with nonalcoholic steatohepatitis. Hepatology 50, 1087–1093 (2009).
Emken, E. A. Metabolism of dietary stearic acid relative to other fatty acids in human subjects. Am. J. Clin. Nutr. 60, 1023S–1028S (1994).
Christinat, N. & Masoodi, M. Comprehensive lipoprotein characterization using lipidomics analysis of human plasma. J. Proteome Res. 16, 2947–2953 (2017).
Kotronen, A. et al. Serum saturated fatty acids containing triacylglycerols are better markers of insulin resistance than total serum triacylglycerol concentrations. Diabetologia 52, 684–690 (2009).
Puri, P. et al. A lipidomic analysis of nonalcoholic fatty liver disease. Hepatology 46, 1081–1090 (2007).
Scorletti, E. & Byrne, C. D. Omega-3 fatty acids, hepatic lipid metabolism, and nonalcoholic fatty liver disease. Annu. Rev. Nutr. 33, 231–248 (2013).
Jeyapal, S. et al. Substitution of linoleic acid with α-linolenic acid or long chain n-3 polyunsaturated fatty acid prevents Western diet induced nonalcoholic steatohepatitis. Sci. Rep. 8, 10953 (2018).
Valenzuela, R. et al. N-3 long-chain polyunsaturated fatty acid supplementation significantly reduces liver oxidative stress in high fat induced steatosis. PLoS ONE 7, e46400 (2012).
Schuster, S. et al. Oxidized linoleic acid metabolites induce liver mitochondrial dysfunction, apoptosis, and NLRP3 activation in mice. J. Lipid Res. 59, 1597–1609 (2018).
Ramsden, C. E. et al. Lowering dietary linoleic acid reduces bioactive oxidized linoleic acid metabolites in humans. Prostaglandins Leukot. Essent. Fat. Acids 87, 135–141 (2012).
Puri, P. et al. The plasma lipidomic signature of nonalcoholic steatohepatitis. Hepatology 50, 1827–1838 (2009).
Loomba, R., Quehenberger, O., Armando, A. & Dennis, E. A. Polyunsaturated fatty acid metabolites as novel lipidomic biomarkers for noninvasive diagnosis of nonalcoholic steatohepatitis. J. Lipid Res. 56, 185–192 (2015).
Caussy, C. et al. Plasma eicosanoids as noninvasive biomarkers of liver fibrosis in patients with nonalcoholic steatohepatitis. Ther. Adv. Gastroenterol. 13, 1756284820923904 (2020).
Feldstein, A. E. et al. Mass spectrometric profiling of oxidized lipid products in human nonalcoholic fatty liver disease and nonalcoholic steatohepatitis. J. Lipid Res. 51, 3046–3054 (2010).
Santoro, N. et al. Oxidized fatty acids: a potential pathogenic link between fatty liver and type 2 diabetes in obese adolescents? Antioxid. Redox Signal. 20, 383–389 (2014).
Zein, C. O. et al. Pentoxifylline decreases oxidized lipid products in nonalcoholic steatohepatitis: new evidence on the potential therapeutic mechanism. Hepatology 56, 1291–1299 (2012).
Musso, G., Gambino, R., Cassader, M., Paschetta, E. & Sircana, A. Specialized proresolving mediators: enhancing nonalcoholic steatohepatitis and fibrosis resolution. Trends Pharmacol. Sci. 39, 387–401 (2018).
Kotronen, A. et al. Hepatic stearoyl-CoA desaturase (SCD)-1 activity and diacylglycerol but not ceramide concentrations are increased in the nonalcoholic human fatty liver. Diabetes 58, 203–208 (2009).
Gorden, D. L. et al. Biomarkers of NAFLD progression: a lipidomics approach to an epidemic. J. Lipid Res. 56, 722–736 (2015).
Valsesia, A., Saris, W. H., Astrup, A., Hager, J. & Masoodi, M. Distinct lipid profiles predict improved glycemic control in obese, nondiabetic patients after a low-caloric diet intervention: the diet, obesity and genes randomized trial. Am. J. Clin. Nutr. 104, 566–575 (2016).
Schwarz, J. M., Linfoot, P., Dare, D. & Aghajanian, K. Hepatic de novo lipogenesis in normoinsulinemic and hyperinsulinemic subjects consuming high-fat, low-carbohydrate and low-fat, high-carbohydrate isoenergetic diets. Am. J. Clin. Nutr. 77, 43–50 (2003).
Mayo, R. et al. Metabolomic-based noninvasive serum test to diagnose nonalcoholic steatohepatitis: results from discovery and validation cohorts. Hepatol. Commun. 2, 807–820 (2018).
Bril, F. et al. Use of a metabolomic approach to non-invasively diagnose non-alcoholic fatty liver disease in patients with type 2 diabetes mellitus. Diabetes Obes. Metab. 20, 1702–1709 (2018).
Oresic, M. et al. Prediction of non-alcoholic fatty-liver disease and liver fat content by serum molecular lipids. Diabetologia 56, 2266–2274 (2013).
Rhee, E. P. et al. Lipid profiling identifies a triacylglycerol signature of insulin resistance and improves diabetes prediction in humans. J. Clin. Invest. 121, 1402–1411 (2011).
Suvitaival, T. et al. Lipidome as a predictive tool in progression to type 2 diabetes in Finnish men. Metabolism 78, 1–12 (2018).
Jorgenrud, B. et al. The influence of sample collection methodology and sample preprocessing on the blood metabolic profile. Bioanalysis 7, 991–1006 (2015).
Ramos-Molina, B. et al. A pilot study of serum sphingomyelin dynamics in subjects with severe obesity and non-alcoholic steatohepatitis after sleeve gastrectomy. Obes. Surg. 29, 983–989 (2019).
Petersen, M. C. & Shulman, G. I. Roles of diacylglycerols and ceramides in hepatic insulin resistance. Trends Pharmacol. Sci. 38, 649–665 (2017).
Pagadala, M., Kasumov, T., McCullough, A. J., Zein, N. N. & Kirwan, J. P. Role of ceramides in nonalcoholic fatty liver disease. Trends Endocrinol. Metab. 23, 365–371 (2012).
Haus, J. M. et al. Plasma ceramides are elevated in obese subjects with type 2 diabetes and correlate with the severity of insulin resistance. Diabetes 58, 337–343 (2009).
Summers, S. A., Chaurasia, B. & Holland, W. L. Metabolic messengers: ceramides. Nat. Metab. 1, 1051–1058 (2019).
Luukkonen, P. K. et al. Hepatic ceramides dissociate steatosis and insulin resistance in patients with non-alcoholic fatty liver disease. J. Hepatol. 64, 1167–1175 (2016).
Apostolopoulou, M. et al. Specific hepatic sphingolipids relate to insulin resistance, oxidative stress, and inflammation in nonalcoholic steatohepatitis. Diabetes Care 41, 1235–1243 (2018).
Chaurasia, B. et al. Targeting a ceramide double bond improves insulin resistance and hepatic steatosis. Science 365, 386–392 (2019).
Xia, J. Y. et al. Targeted induction of ceramide degradation leads to improved systemic metabolism and reduced hepatic steatosis. Cell Metab. 22, 266–278 (2015).
Hyysalo, J. et al. Circulating triacylglycerol signatures in nonalcoholic fatty liver disease associated with the I148M variant in PNPLA3 and with obesity. Diabetes 63, 312–322 (2014).
Luukkonen, P. K. et al. Human PNPLA3-I148M variant increases hepatic retention of polyunsaturated fatty acids. JCI Insight 4, e127902 (2019).
Luukkonen, P. K. et al. Impaired hepatic lipid synthesis from polyunsaturated fatty acids in TM6SF2 E167K variant carriers with NAFLD. J. Hepatol. 67, 128–136 (2017).
Zhou, Y. et al. Circulating triacylglycerol signatures and insulin sensitivity in NAFLD associated with the E167K variant in TM6SF2. J. Hepatol. 62, 657–663 (2015).
Mancina, R. M. et al. The MBOAT7-TMC4 variant rs641738 increases risk of nonalcoholic fatty liver disease in individuals of European descent. Gastroenterology 150, 1219–1230.e6 (2016).
Lee, H. C. et al. Caenorhabditis elegans mboa-7, a member of the MBOAT family, is required for selective incorporation of polyunsaturated fatty acids into phosphatidylinositol. Mol. Biol. Cell 19, 1174–1184 (2008).
Meroni, M. et al. Mboat7 down-regulation by hyper-insulinemia induces fat accumulation in hepatocytes. EBioMedicine 52, 102658 (2020).
Helsley, R. N. et al. Obesity-linked suppression of membrane-bound O-acyltransferase 7 (MBOAT7) drives non-alcoholic fatty liver disease. Elife 8, e49882 (2019).
Tanaka, Y. et al. LPIAT1/MBOAT7 depletion increases triglyceride synthesis fueled by high phosphatidylinositol turnover. Gut 70, 180–193 (2021).
Fondevila, M. F. et al. The L-α-lysophosphatidylinositol/G protein-coupled receptor 55 system induces the development of nonalcoholic steatosis and steatohepatitis. Hepatology 73, 606–624 (2021).
Luukkonen, P. K. et al. Hydroxysteroid 17-β dehydrogenase 13 variant increases phospholipids and protects against fibrosis in nonalcoholic fatty liver disease. JCI Insight 5, e132158 (2020).
Luukkonen, P. K. et al. MARC1 variant rs2642438 increases hepatic phosphatidylcholines and decreases severity of non-alcoholic fatty liver disease in humans. J. Hepatol. 73, 725–726 (2020).
Chiang, J. Y. Regulation of bile acid synthesis: pathways, nuclear receptors, and mechanisms. J. Hepatol. 40, 539–551 (2004).
Behr, A. C., Plinsch, C., Braeuning, A. & Buhrke, T. Activation of human nuclear receptors by perfluoroalkylated substances (PFAS). Toxicol. Vitro 62, 104700 (2020).
Bjork, J. A., Butenhoff, J. L. & Wallace, K. B. Multiplicity of nuclear receptor activation by PFOA and PFOS in primary human and rodent hepatocytes. Toxicology 288, 8–17 (2011).
Zhang, L. et al. Persistent organic pollutants modify gut microbiota-host metabolic homeostasis in mice through aryl hydrocarbon receptor activation. Environ. Health Perspect. 123, 679–688 (2015).
Chiang, J. Y. Recent advances in understanding bile acid homeostasis. F1000Res 6, 2029 (2017).
Honda, A. et al. Regulation of bile acid metabolism in mouse models with hydrophobic bile acid composition. J. Lipid Res. 61, 54–69 (2020).
Ticho, A. L., Malhotra, P., Dudeja, P. K., Gill, R. K. & Alrefai, W. A. Bile acid receptors and gastrointestinal functions. Liver Res. 3, 31–39 (2019).
Jiao, N. et al. Suppressed hepatic bile acid signalling despite elevated production of primary and secondary bile acids in NAFLD. Gut 67, 1881–1891 (2018).
Pineda Torra, I. et al. Bile acids induce the expression of the human peroxisome proliferator-activated receptor α gene via activation of the farnesoid X receptor. Mol. Endocrinol. 17, 259–272 (2003).
Kast, H. R. et al. Farnesoid X-activated receptor induces apolipoprotein C-II transcription: a molecular mechanism linking plasma triglyceride levels to bile acids. Mol. Endocrinol. 15, 1720–1728 (2001).
Dasarathy, S. et al. Elevated hepatic fatty acid oxidation, high plasma fibroblast growth factor 21, and fasting bile acids in nonalcoholic steatohepatitis. Eur. J. Gastroenterol. Hepatol. 23, 382–388 (2011).
Aranha, M. M. et al. Bile acid levels are increased in the liver of patients with steatohepatitis. Eur. J. Gastroenterol. Hepatol. 20, 519–525 (2008).
Mouzaki, M. et al. Bile acids and dysbiosis in non-alcoholic fatty liver disease. PLoS ONE 11, e0151829 (2016).
Puri, P. et al. The presence and severity of nonalcoholic steatohepatitis is associated with specific changes in circulating bile acids. Hepatology 67, 534–548 (2018).
Christinat, N., Valsesia, A. & Masoodi, M. Untargeted profiling of bile acids and lysophospholipids identifies the lipid signature associated with glycemic outcome in an obese non-diabetic clinical cohort. Biomolecules 10, 1049 (2020).
Wruck, W. & Adjaye, J. Meta-analysis reveals up-regulation of cholesterol processes in non-alcoholic and down-regulation in alcoholic fatty liver disease. World J. Hepatol. 9, 443–454 (2017).
Legry, V. et al. Bile acid alterations are associated with insulin resistance, but not with NASH, in obese subjects. J. Clin. Endocrinol. Metab. 102, 3783–3794 (2017).
Bechmann, L. P. et al. Free fatty acids repress small heterodimer partner (SHP) activation and adiponectin counteracts bile acid-induced liver injury in superobese patients with nonalcoholic steatohepatitis. Hepatology 57, 1394–1406 (2013).
Lake, A. D. et al. Decreased hepatotoxic bile acid composition and altered synthesis in progressive human nonalcoholic fatty liver disease. Toxicol. Appl. Pharmacol. 268, 132–140 (2013).
Tanaka, N., Matsubara, T., Krausz, K. W., Patterson, A. D. & Gonzalez, F. J. Disruption of phospholipid and bile acid homeostasis in mice with nonalcoholic steatohepatitis. Hepatology 56, 118–129 (2012).
Valanejad, L. et al. Dysregulation of Δ(4)-3-oxosteroid 5β-reductase in diabetic patients: implications and mechanisms. Mol. Cell Endocrinol. 470, 127–141 (2018).
Chen, J. et al. Ratio of conjugated chenodeoxycholic to muricholic acids is associated with severity of nonalcoholic steatohepatitis. Obesity 27, 2055–2066 (2019).
Chavez-Talavera, O., Tailleux, A., Lefebvre, P. & Staels, B. Bile acid control of metabolism and inflammation in obesity, type 2 diabetes, dyslipidemia, and nonalcoholic fatty liver disease. Gastroenterology 152, 1679–1694.e3 (2017).
Haeusler, R. A., Astiarraga, B., Camastra, S., Accili, D. & Ferrannini, E. Human insulin resistance is associated with increased plasma levels of 12α-hydroxylated bile acids. Diabetes 62, 4184–4191 (2013).
Aragones, G. et al. Circulating microbiota-derived metabolites: a “liquid biopsy? Int. J. Obes. 44, 875–885 (2020).
Ferslew, B. C. et al. Altered bile acid metabolome in patients with nonalcoholic steatohepatitis. Dig. Dis. Sci. 60, 3318–3328 (2015).
Aron-Wisnewsky, J. et al. Gut microbiota and human NAFLD: disentangling microbial signatures from metabolic disorders. Nat. Rev. Gastroenterol. Hepatol. 17, 279–297 (2020).
Lefere, S. & Tacke, F. Macrophages in obesity and non-alcoholic fatty liver disease: crosstalk with metabolism. JHEP Rep. 1, 30–43 (2019).
Szabo, G., Bala, S., Petrasek, J. & Gattu, A. Gut-liver axis and sensing microbes. Dig. Dis. 28, 737–744 (2010).
Ji, Y., Yin, Y., Li, Z. & Zhang, W. Gut microbiota-derived components and metabolites in the progression of non-alcoholic fatty liver disease (NAFLD). Nutrients 11, 1712 (2019).
Jasirwan, C. O. M., Lesmana, C. R. A., Hasan, I., Sulaiman, A. S. & Gani, R. A. The role of gut microbiota in non-alcoholic fatty liver disease: pathways of mechanisms. Biosci. Microbiota Food Health 38, 81–88 (2019).
Liu, Q. et al. Role and effective therapeutic target of gut microbiota in NAFLD/NASH. Exp. Ther. Med. 18, 1935–1944 (2019).
Pan, X., Wen, S. W., Kaminga, A. C. & Liu, A. Gut metabolites and inflammation factors in non-alcoholic fatty liver disease: a systematic review and meta-analysis. Sci. Rep. 10, 8848 (2020).
Jiang, W. et al. Dysbiosis gut microbiota associated with inflammation and impaired mucosal immune function in intestine of humans with non-alcoholic fatty liver disease. Sci. Rep. 5, 8096 (2015).
Mouzaki, M. et al. Intestinal microbiota in patients with nonalcoholic fatty liver disease. Hepatology 58, 120–127 (2013).
Rau, M. et al. Fecal SCFAs and SCFA-producing bacteria in gut microbiome of human NAFLD as a putative link to systemic T-cell activation and advanced disease. United Eur. Gastroenterol. J. 6, 1496–1507 (2018).
Ramirez-Perez, O., Cruz-Ramon, V., Chinchilla-Lopez, P. & Mendez-Sanchez, N. The role of the gut microbiota in bile acid metabolism. Ann. Hepatol. 16, s15–s20 (2017).
van Best, N. et al. Bile acids drive the newborn’s gut microbiota maturation. Nat Commun 11, 3692 (2020).
Tremblay, S. et al. Bile acid administration elicits an intestinal antimicrobial program and reduces the bacterial burden in two mouse models of enteric infection. Infect. Immun. 85, e00942 (2017).
Ridlon, J. M., Kang, D. J. & Hylemon, P. B. Bile salt biotransformations by human intestinal bacteria. J. Lipid Res. 47, 241–259 (2006).
Sarathy, J. et al. The Yin and Yang of bile acid action on tight junctions in a model colonic epithelium. Physiol. Rep. 5, e13294 (2017).
Corbin, K. D. & Zeisel, S. H. Choline metabolism provides novel insights into nonalcoholic fatty liver disease and its progression. Curr. Opin. Gastroenterol. 28, 159–165 (2012).
Chen, Y. M. et al. Associations of gut-flora-dependent metabolite trimethylamine-N-oxide, betaine and choline with non-alcoholic fatty liver disease in adults. Sci. Rep. 6, 19076 (2016).
Hoyles, L. et al. Molecular phenomics and metagenomics of hepatic steatosis in non-diabetic obese women. Nat. Med. 24, 1070–1080 (2018).
Caussy, C. & Loomba, R. Gut microbiome, microbial metabolites and the development of NAFLD. Nat. Rev. Gastroenterol. Hepatol. 15, 719–720 (2018).
Krishnan, S. et al. Gut microbiota-derived tryptophan metabolites modulate inflammatory response in hepatocytes and macrophages. Cell Rep. 23, 1099–1111 (2018).
Gastaldelli, A. & Cusi, K. From NASH to diabetes and from diabetes to NASH: mechanisms and treatment options. JHEP Rep. 1, 312–328 (2019).
Wolfe, R. R. & Chinkes, D. L. Isotope Tracers in Metabolic Research: Principles and Practice of Kinetic Analysis (Wiley, 2004).
Cusi, K., Kashyap, S., Gastaldelli, A., Bajaj, M. & Cersosimo, E. Effects on insulin secretion and insulin action of a 48-h reduction of plasma free fatty acids with acipimox in nondiabetic subjects genetically predisposed to type 2 diabetes. Am. J. Physiol. Endocrinol. Metab. 292, E1775–E1781 (2007).
Smith, G. I. et al. Insulin resistance drives hepatic de novo lipogenesis in nonalcoholic fatty liver disease. J. Clin. Invest. 130, 1453–1460 (2020).
Kalhan, S. C., Bugianesi, E., McCullough, A. J., Hanson, R. W. & Kelley, D. E. Estimates of hepatic glyceroneogenesis in type 2 diabetes mellitus in humans. Metabolism 57, 305–312 (2008).
Jin, E. S., Browning, J. D., Murphy, R. E. & Malloy, C. R. Fatty liver disrupts glycerol metabolism in gluconeogenic and lipogenic pathways in humans. J. Lipid Res. 59, 1685–1694 (2018).
Lytle, K. A. et al. Hepatic fatty acid balance and hepatic fat content in humans with severe obesity. J. Clin. Endocrinol. Metab. 104, 6171–6181 (2019).
Satapati, S. et al. Mitochondrial metabolism mediates oxidative stress and inflammation in fatty liver. J. Clin. Invest. 125, 4447–4462 (2015).
Sunny, N. E. et al. Cross-talk between branched-chain amino acids and hepatic mitochondria is compromised in nonalcoholic fatty liver disease. Am. J. Physiol. Endocrinol. Metab. 309, E311–E319 (2015).
Burla, B. et al. MS-based lipidomics of human blood plasma: a community-initiated position paper to develop accepted guidelines. J. Lipid Res. 59, 2001–2017 (2018).
Bowden, J. A. et al. Harmonizing lipidomics: NIST interlaboratory comparison exercise for lipidomics using SRM 1950-metabolites in frozen human plasma. J. Lipid Res. 58, 2275–2288 (2017).
Feldman, A. et al. Clinical and metabolic characterization of lean Caucasian subjects with non-alcoholic fatty liver. Am. J. Gastroenterol. 112, 102–110 (2017).
Pirola, C. J. & Sookoian, S. Multiomics biomarkers for the prediction of nonalcoholic fatty liver disease severity. World J. Gastroenterol. 24, 1601–1615 (2018).
Sookoian, S. & Pirola, C. J. Liver enzymes, metabolomics and genome-wide association studies: from systems biology to the personalized medicine. World J. Gastroenterol. 21, 711–725 (2015).
Valsesia, A. et al. Integrative phenotyping of glycemic responders upon clinical weight loss using multi-omics. Sci. Rep. 10, 9236 (2020).
Magnusdottir, S. et al. Generation of genome-scale metabolic reconstructions for 773 members of the human gut microbiota. Nat. Biotechnol. 35, 81–89 (2017).
Sen, P. & Oresic, M. Metabolic modeling of human gut microbiota on a genome scale: an overview. Metabolites 9, 22 (2019).
Gastaldelli A. in Non-Alcoholic Fatty Liver Disease (ed. Bugianesi, E.) 49–71 (Springer, 2020)
Kleiner, D. E. et al. Design and validation of a histological scoring system for nonalcoholic fatty liver disease. Hepatology 41, 1313–1321 (2005).
Acknowledgements
All authors of this manuscript are members of the Liver Investigation: Testing Marker Utility in Steatohepatitis (LITMUS) Consortium. The authors are supported by the EU’s Horizon 2020 Programme under the EPoS project (grant no. 634413) and the Innovative Medicines Initiative 2 Joint Undertaking under the LITMUS project (grant no. 777377) and Swiss National Science Foundation (SNSF) (grant no. 190686). This Joint Undertaking receives support from the EU’s Horizon 2020 programme and EFPIA.
Author information
Authors and Affiliations
Contributions
M.M. researched data for the article, made a substantial contribution to discussion of content, wrote the first draft, and reviewed/edited the manuscript before submission. A.G. and T.H. made a substantial contribution to discussion of content, wrote the article, and reviewed/edited the manuscript before submission. E.A. researched data for the article and wrote the article. C.A. and M.G. researched data for the article, and reviewed/edited the manuscript before submission. J.B., Q.M.A., O.M., P.O. and J.M.M. reviewed/edited the manuscript before submission. J.-F.D. made a substantial contribution to discussion of content, and reviewed/edited the manuscript before submission. M.O. researched data for the article, made a substantial contribution to discussion of content, contributed to writing and reviewed/edited the manuscript before submission.
Corresponding authors
Ethics declarations
Competing interests
J.B. was an employee and shareholder of Pfizer. E.A., C.A. and P.O. are employees of OWL Metabolomics. A.G. is a consultant for Eli Lilly, Inventiva, Genentech, Menarini, Gilead, Novo Nordisk, AstraZeneca and Boehringer outside the submitted work. J.-F.D. is on advisory committees for Abbvie, Bayer, BMS, Falk, Genfit, Genkyotex, Gilead Science, HepaRegenix, Intercept, Lilly, Merck and Novartis; speaking and teaching at Bayer, BMS, Intercept, Genfit, Gilead Science and Novartis. Q.M.A. has active research collaborations (including research collaborations supported through the EU IMI2 LITMUS Consortium*) with Abbvie, Antaros Medical*, Allergan/Tobira*, AstraZeneca*, BMS*, Boehringer Ingelheim International GMBH*, Echosens*, Ellegaard Gottingen Minipigs AS*, Eli Lilly & Company Ltd.*, Exalenz Bioscience Ltd.*, Genfit SA*, Glympse Bio, GlaxoSmithKline, HistoIndex*, Intercept Pharma Europe Ltd.*, iXscient Ltd.*, Nordic Bioscience*, Novartis Pharma AG*, Novo Nordisk A/S*, One Way Liver SL*, Perspectum Diagnostics*, Pfizer Ltd.*, Resoundant*, Sanofi-Aventis Deutschland GMBH*, SomaLogic Inc.* and Takeda Pharmaceuticals International SA*, acts as a consultant for 89Bio, Abbott Laboratories, Acuitas Medical, Allergan/Tobira, Altimmune, AstraZeneca, Axcella, Blade, BMS, BNN Cardio, Celgene, Cirius, CymaBay, EcoR1, E3Bio, Eli Lilly & Company Ltd., Galmed, Genentech, Genfit SA, Gilead, Grunthal, HistoIndex, Indalo, Imperial Innovations, Intercept Pharma Europe Ltd., Inventiva, IQVIA, Janssen, Madrigal, MedImmune, Metacrine, NewGene, NGMBio, North Sea Therapeutics, Novartis, Novo Nordisk A/S, Pfizer Ltd., Poxel, ProSciento, Raptor Pharma, Servier, Terns, Viking Therapeutics and Speaker for Abbott Laboratories, Allergan/Tobira, BMS, Clinical Care Options, Falk, Fishawack, Genfit SA, Gilead, Integritas Communications, Kenes and MedScape, and has received royalties from Elsevier Ltd (Davidson’s Principles & Practice of Medicine textbook). All other authors declare no competing interests.
Additional information
Peer review information
Nature Reviews Gastroenterology & Hepatology thanks C. Pirola, F. Bril and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Related links
IMI2 LITMUS: https://litmus-project.eu/
Rights and permissions
About this article
Cite this article
Masoodi, M., Gastaldelli, A., Hyötyläinen, T. et al. Metabolomics and lipidomics in NAFLD: biomarkers and non-invasive diagnostic tests. Nat Rev Gastroenterol Hepatol 18, 835–856 (2021). https://doi.org/10.1038/s41575-021-00502-9
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41575-021-00502-9
This article is cited by
-
Glutamine prevents high-fat diet-induced hepatic lipid accumulation in mice by modulating lipolysis and oxidative stress
Nutrition & Metabolism (2024)
-
Development of non-alcoholic steatohepatitis is associated with gut microbiota but not with oxysterol enzymes CH25H, EBI2, or CYP7B1 in mice
BMC Microbiology (2024)
-
Transcriptomics-driven metabolic pathway analysis reveals similar alterations in lipid metabolism in mouse MASH model and human
Communications Medicine (2024)
-
Muscular lipidomics and transcriptomics reveal the effects of bile acids on lipid metabolism in high-fat diet-fed grouper
Fish Physiology and Biochemistry (2024)
-
Sex differences in the association between adipose insulin resistance and non-alcoholic fatty liver disease in Chinese adults
Biology of Sex Differences (2023)