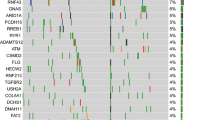
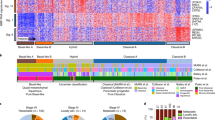
Abstract
Pancreatic cancer is a genetic disease, and the recurrent genetic alterations characteristic of pancreatic cancer indicate the cellular processes that are targeted for malignant transformation. In addition to somatic alterations in the most common driver genes (KRAS, CDKN2A, TP53 and SMAD4), large-scale studies have revealed major roles for genetic alterations of the SWI/SNF and COMPASS complexes, copy number alterations in GATA6 and MYC that partially define phenotypes of pancreatic cancer, and the role(s) of polyploidy and chromothripsis as factors contributing to pancreatic cancer biology and progression. Germline variants that increase the risk of pancreatic cancer continue to be discovered along with a greater appreciation of the features of pancreatic cancers with mismatch repair deficiencies and homologous recombination deficiencies that confer sensitivity to therapeutic targeting. Wild-type KRAS pancreatic cancers, some of which are driven by alternative oncogenic events affecting NRG1 or NTRK1 — for which targeted therapies exist — further underscore that pancreatic cancer is formally entering the era of precision medicine. Given the vast developments within this field, here we review the wide-ranging and most current information related to pancreatic cancer genomics with the goal of integrating this information into a unifying description of the life history of pancreatic cancer.
Key Points
-
The natural history of pancreatic cancer is characterized by both genetic and epigenetic alterations that contribute to its formation, progression and resistance to therapy.
-
Most pancreatic cancers arise due to the accumulation of somatic alterations in a recurrent set of genes; however, some patients might develop pancreatic cancer owing to a genetic predisposition.
-
Rare subsets of pancreatic cancers arise in association with a genetic alteration that is targetable.
-
The pancreatic cancer stroma, inclusive of the immune system, acts as a dynamic selective pressure to which the neoplasm continuously adapts.
-
Distinct genomic events are associated with pancreatic cancer phenotypes that are differentially sensitive to currently available therapies.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Rawla, P., Sunkara, T. & Gaduputi, V. Epidemiology of pancreatic cancer: global trends, etiology and risk factors. World J. Oncol. 10, 10–27 (2019).
Grossberg, A. J. et al. Multidisciplinary standards of care and recent progress in pancreatic ductal adenocarcinoma. CA Cancer J. Clin. 70, 375–403 (2020).
Conroy, T. et al. FOLFIRINOX or gemcitabine as adjuvant therapy for pancreatic cancer. N. Engl. J. Med. 379, 2395–2406 (2018).
Von Hoff, D. D. et al. Increased survival in pancreatic cancer with nab-paclitaxel plus gemcitabine. N. Engl. J. Med. 369, 1691–1703 (2013).
Hendifar, A. et al. Influence of body mass index and albumin on perioperative morbidity and clinical outcomes in resected pancreatic adenocarcinoma. PLoS ONE 11, e0152172 (2016).
Lomberk, G., Dusetti, N., Iovanna, J. & Urrutia, R. Emerging epigenomic landscapes of pancreatic cancer in the era of precision medicine. Nat. Commun. 10, 3875 (2019).
Makohon-Moore, A. & Iacobuzio-Donahue, C. A. Pancreatic cancer biology and genetics from an evolutionary perspective. Nat. Rev. Cancer 16, 553–565 (2016).
Jones, S. et al. Core signaling pathways in human pancreatic cancers revealed by global genomic analyses. Science 321, 1801–1806 (2008).
Biankin, A. V. et al. Pancreatic cancer genomes reveal aberrations in axon guidance pathway genes. Nature 491, 399–405 (2012).
Witkiewicz, A. K. et al. Whole-exome sequencing of pancreatic cancer defines genetic diversity and therapeutic targets. Nat. Commun. 6, 6744 (2015).
Raphael, B. J. et al. Integrated genomic characterization of pancreatic ductal adenocarcinoma. Cancer Cell 32, 185–203 (2017).
Waddell, N. et al. Whole genomes redefine the mutational landscape of pancreatic cancer. Nature 518, 495–501 (2015).
Notta, F. et al. A renewed model of pancreatic cancer evolution based on genomic rearrangement patterns. Nature 538, 378–382 (2016).
Connor, A. A. et al. Integration of genomic and transcriptional features in pancreatic cancer reveals increased cell cycle progression in metastases. Cancer Cell 35, 267–282 (2019).
Hansel, D. E., Kern, S. E. & Hruban, R. H. Molecular pathogenesis of pancreatic cancer. Annu. Rev. Genomics Hum. Genet. 4, 237–256 (2003).
Waters, A. M. & Der, C. J. KRAS: the critical driver and therapeutic target for pancreatic cancer. Cold Spring Harb. Perspect. Med. 8, a031435 (2018).
Ruscetti, M. et al. Senescence-induced vascular remodeling creates therapeutic vulnerabilities in pancreas cancer. Cell 181, 424–441 (2020).
Collisson, E. A. et al. Subtypes of pancreatic ductal adenocarcinoma and their differing responses to therapy. Nat. Med. 17, 500–503 (2011).
Moffitt, R. A. et al. Virtual microdissection identifies distinct tumor- and stroma-specific subtypes of pancreatic ductal adenocarcinoma. Nat. Genet. 47, 1168–1178 (2015).
Bailey, P. et al. Genomic analyses identify molecular subtypes of pancreatic cancer. Nature 531, 47–52 (2016).
Chan-Seng-Yue, M. et al. Transcription phenotypes of pancreatic cancer are driven by genomic events during tumor evolution. Nat. Genet. 52, 231–240 (2020).
Hayashi, A. et al. A unifying paradigm for transcriptional heterogeneity and squamous features in pancreatic ductal adenocarcinoma. Nat. Cancer 1, 59–74 (2020).
O’Reilly, E. M. & Hechtman, J. F. Tumour response to TRK inhibition in a patient with pancreatic adenocarcinoma harbouring an NTRK gene fusion. Ann. Oncol. 30, VIII36–VIII40 (2019).
Hayashi, A. et al. Evolutionary dynamics of non-coding regions in pancreatic ductal adenocarcinoma. Preprint at bioRxiv https://doi.org/10.1101/2020.09.11.294389 (2020).
Tokheim, C. & Karchin, R. CHASMplus reveals the scope of somatic missense mutations driving human cancers. Cell Syst. 9, 9–23 (2019).
Tokheim, C. J., Papadopoulos, N., Kinzler, K. W., Vogelstein, B. & Karchin, R. Evaluating the evaluation of cancer driver genes. Proc. Natl Acad. Sci. USA 113, 14330–14335 (2016).
Vogelstein, B. et al. Cancer genome landscapes. Science 340, 1546–1558 (2013).
Garraway, L. A. & Lander, E. S. Lessons from the cancer genome. Cell 153, 17–37 (2013).
Reiter, J. G. et al. Minimal functional driver gene heterogeneity among untreated metastases. Science 361, 1033–1037 (2018).
Rheinbay, E. et al. Analyses of non-coding somatic drivers in 2,658 cancer whole genomes. Nature 578, 102–111 (2020).
Shuai, S. et al. Combined burden and functional impact tests for cancer driver discovery using DriverPower. Nat. Commun. 11, 734 (2020).
Chakravarty, D. et al. OncoKB: a precision oncology knowledge base. JCO Precis. Oncol. 2017, PO.17.00011 (2017).
Kurosaki, T., Popp, M. W. & Maquat, L. E. Quality and quantity control of gene expression by nonsense-mediated mRNA decay. Nat. Rev. Mol. Cell Biol. 20, 406–420 (2019).
Cherry, S. & Lynch, K. W. Alternative splicing and cancer: insights, opportunities, and challenges from an expanding view of the transcriptome. Genes Dev. 34, 1005–1016 (2020).
Luksza, M. et al. A neoantigen fitness model predicts tumour response to checkpoint blockade immunotherapy. Nature 551, 517–520 (2017).
Alexandrov, L. B. et al. Signatures of mutational processes in human cancer. Nature 500, 415–421 (2013).
Balachandran, V. P. et al. Identification of unique neoantigen qualities in long-term survivors of pancreatic cancer. Nature 551, S12–S16 (2017).
Kryklyva, V. et al. Medullary pancreatic carcinoma due to somatic POLE mutation: a distinctive pancreatic carcinoma with marked long-term survival. Pancreas 49, 999–1003 (2020).
Humphris, J. L. et al. Hypermutation in pancreatic cancer. Gastroenterology 152, 68–74 (2017).
Le, D. T. et al. PD-1 blockade in tumors with mismatch-repair deficiency. N. Engl. J. Med. 372, 2509–2520 (2015).
Feigin, M. E. et al. Recurrent noncoding regulatory mutations in pancreatic ductal adenocarcinoma. Nat. Genet. 49, 825–833 (2017).
Whittle, M. C. et al. RUNX3 controls a metastatic switch in pancreatic ductal adenocarcinoma. Cell 161, 1345–1360 (2015).
Karmakar, S. et al. MicroRNA regulation of K-Ras in pancreatic cancer and opportunities for therapeutic intervention. Semin. Cancer Biol. 54, 63–71 (2019).
Wu, J. et al. Whole-exome sequencing of neoplastic cysts of the pancreas reveals recurrent mutations in components of ubiquitin-dependent pathways. Proc. Natl Acad. Sci. USA 108, 21188–21193 (2011).
Bozic, I. et al. Accumulation of driver and passenger mutations during tumor progression. Proc. Natl Acad. Sci. USA 107, 18545–18550 (2010).
Alexandrov, L. B. et al. The repertoire of mutational signatures in human cancer. Nature 578, 94–101 (2020).
Connor, A. A. et al. Association of distinct mutational signatures with correlates of increased immune activity in pancreatic ductal adenocarcinoma. JAMA Oncol. 3, 774–783 (2017).
Sakamoto, H. et al. The evolutionary origins of recurrent pancreatic cancer. Cancer Discov. 10, 792–805 (2020).
Iodice, S., Gandini, S., Maisonneuve, P. & Lowenfels, A. B. Tobacco and the risk of pancreatic cancer: a review and meta-analysis. Langenbeck’s Arch. Surg. 393, 535–545 (2008).
Lynch, S. M. et al. Cigarette smoking and pancreatic cancer: a pooled analysis from the Pancreatic Cancer Cohort Consortium. Am. J. Epidemiol. 170, 403–413 (2009).
Hu, Z. I. et al. Evaluating mismatch repair deficiency in pancreatic adenocarcinoma: challenges and recommendations. Clin. Cancer Res. 24, 1326–1336 (2018).
Helleday, T., Eshtad, S. & Nik-Zainal, S. Mechanisms underlying mutational signatures in human cancers. Nat. Rev. Genet. 15, 585–598 (2014).
Suspène, R. et al. Somatic hypermutation of human mitochondrial and nuclear DNA by APOBEC3 cytidine deaminases, a pathway for DNA catabolism. Proc. Natl Acad. Sci. USA 108, 4858–4863 (2011).
Landry, S., Narvaiza, I., Linfesty, D. C. & Weitzman, M. D. APOBEC3A can activate the DNA damage response and cause cell-cycle arrest. EMBO Rep. 12, 444–450 (2011).
Campbell, P. J. et al. Pan-cancer analysis of whole genomes. Nature 578, 82–93 (2020).
Evans, M. D., Dizdaroglu, M. & Cooke, M. S. Oxidative DNA damage and disease: induction, repair and significance. Mutat. Res. Rev. Mutat. Res. 567, 1–61 (2004).
Al-Tassan, N. et al. Inherited variants of MYH associated with somatic G:C→T:A mutations in colorectal tumors. Nat. Genet. 30, 227–232 (2002).
Georgeson, P. et al. Evaluating the utility of tumour mutational signatures for identifying hereditary colorectal cancer and polyposis syndrome carriers. Gut https://doi.org/10.1136/gutjnl-2019-320462 (2021).
Bielski, C. M. et al. Genome doubling shapes the evolution and prognosis of advanced cancers. Nat. Genet. 50, 1189–1195 (2018).
Stephens, P. J. et al. Massive genomic rearrangement acquired in a single catastrophic event during cancer development. Cell 144, 27–40 (2011).
Cortés-Ciriano, I. et al. Comprehensive analysis of chromothripsis in 2,658 human cancers using whole-genome sequencing. Nat. Genet. 52, 331–341 (2020).
Maley, C. C. et al. Classifying the evolutionary and ecological features of neoplasms. Nat. Rev. Cancer 17, 605–619 (2017).
Reiter, J. G. & Iacobuzio-Donahue, C. A. Pancreatic cancer: pancreatic carcinogenesis–several small steps or one giant leap? Nat. Rev. Gastroenterol. Hepatol. 14, 7–8 (2017).
Storz, P. & Crawford, H. C. Carcinogenesis of pancreatic ductal adenocarcinoma. Gastroenterology 158, 2072–2081 (2020).
Buscail, L., Bournet, B. & Cordelier, P. Role of oncogenic KRAS in the diagnosis, prognosis and treatment of pancreatic cancer. Nat. Rev. Gastroenterol. Hepatol. 17, 153–168 (2020).
Hart, P. A. et al. Type 3c (pancreatogenic) diabetes mellitus secondary to chronic pancreatitis and pancreatic cancer. Lancet Gastroenterol. Hepatol. 1, 226–237 (2016).
Yadav, D. et al. Prospective Evaluation of Chronic Pancreatitis for Epidemiologic and Translational Studies: rationale and study design for PROCEED from the Consortium for the Study of Chronic Pancreatitis, Diabetes, and Pancreatic Cancer. Pancreas 47, 1229–1238 (2018).
Grabocka, E. & Bar-Sagi, D. Mutant KRAS enhances tumor cell fitness by upregulating stress granules. Cell 167, 1803–1813 (2016).
Zhao, Y. et al. Oncogene-induced senescence limits the progression of pancreatic neoplasia through production of activin A. Cancer Res. 80, 3359–3371 (2020).
Commisso, C. et al. Macropinocytosis of protein is an amino acid supply route in Ras-transformed cells. Nature 497, 633–637 (2013).
Shi, C. et al. KRAS2 mutations in human pancreatic acinar-ductal metaplastic lesions are limited to those with PanIN: implications for the human pancreatic cancer cell of origin. Mol. Cancer Res. 7, 230–236 (2009).
Kanda, M. et al. Presence of somatic mutations in most early-stage pancreatic intraepithelial neoplasia. Gastroenterology 142, 730–733 (2012).
Burgess, M. R. et al. KRAS allelic imbalance enhances fitness and modulates MAP kinase dependence in cancer. Cell 168, 817–829 (2017).
Mueller, S. et al. Evolutionary routes and KRAS dosage define pancreatic cancer phenotypes. Nature 554, 62–68 (2018).
Heining, C. et al. NRG1 fusions in KRAS wild-type pancreatic cancer. Cancer Discov. 8, 1087–1095 (2018).
Jones, M. R. et al. NRG1 gene fusions are recurrent, clinically actionable gene rearrangements in KRAS wild-type pancreatic ductal adenocarcinoma. Clin. Cancer Res. 25, 4674–4681 (2019).
Gil, J. & Peters, G. Regulation of the INK4b-ARF-INK4a tumour suppressor locus: all for one or one for all. Nat. Rev. Mol. Cell Biol. 7, 667–677 (2006).
Kim, W. Y. & Sharpless, N. E. The regulation of INK4/ARF in cancer and aging. Cell 127, 265–275 (2006).
Schutte, M. et al. Abrogation of the Rb/p16 tumor-suppressive pathway in virtually all pancreatic carcinomas. Cancer Res. 57, 3126–3130 (1997).
Yachida, S. et al. Clinical significance of the genetic landscape of pancreatic cancer and implications for identification of potential long-term survivors. Clin. Cancer Res. 18, 6339–6347 (2012).
Klein, W. M., Hruban, R. H., Klein-Szanto, A. J. P. & Wilentz, R. E. Direct correlation between proliferative activity and dysplasia in pancreatic intraepithelial neoplasia (panIN): additional evidence for a recently proposed model of progression. Mod. Pathol. 15, 441–447 (2002).
Yachida, S. et al. Distant metastasis occurs late during the genetic evolution of pancreatic cancer. Nature 467, 1114–1117 (2010).
Hosoda, W. et al. Genetic analyses of isolated high-grade pancreatic intraepithelial neoplasia (HG-PanIN) reveal paucity of alterations in TP53 and SMAD4. J. Pathol. 242, 16–23 (2017).
Makohon-Moore, A. P. et al. Precancerous neoplastic cells can move through the pancreatic ductal system. Nature 561, 201–205 (2018).
Vogelstein, B., Lane, D. & Levine, A. J. Surfing the p53 network. Nature 408, 307–310 (2000).
Morton, J. P. et al. Mutant p53 drives metastasis and overcomes growth arrest/senescence in pancreatic cancer. Proc. Natl Acad. Sci. USA 107, 246–251 (2010).
Morton, J. P., Klimstra, D. S., Mongeau, M. E. & Lewis, B. C. Trp53 deletion stimulates the formation of metastatic pancreatic tumors. Am. J. Pathol. 172, 1081–1087 (2008).
Hayashi, A. et al. Genetic and clinical correlates of entosis in pancreatic ductal adenocarcinoma. Mod. Pathol. 33, 1822–1831 (2020).
Florey, O., Kim, S. & Overholtzer, M. Entosis: cell-in-cell formation that kills through entotic cell death. Curr. Mol. Med. 15, 861–866 (2015).
Shu, Z., Row, S. & Deng, W. M. Endoreplication: the good, the bad, and the ugly. Trends Cell Biol. 28, 465–474 (2018).
David, C. J. & Massagué, J. Contextual determinants of TGFβ action in development, immunity and cancer. Nat. Rev. Mol. Cell Biol. 19, 419–435 (2018).
Hahn, S. A. et al. DPC4, a candidate tumor suppressor gene at human chromosome 18q21.1. Science 271, 350–353 (1996).
Togashi, Y. et al. Homozygous deletion of the activin A receptor, type IB gene is associated with an aggressive cancer phenotype in pancreatic cancer. Mol. Cancer 13, 126 (2014).
Su, G. H. et al. ACVR1B (ALK4, activin receptor type 1B) gene mutations in pancreatic carcinoma. Proc. Natl Acad. Sci. USA 98, 3254–3257 (2001).
Hempen, P. M. et al. Evidence of selection for clones having genetic inactivation of the activin A type II receptor (ACVR2) gene in gastrointestinal cancers. Cancer Res. 63, 994–999 (2003).
Ryu, B. & Kern, S. E. The essential similarity of TGFβ and activin receptor transcriptional responses in cancer cells. Cancer Biol. Ther. 2, 164–170 (2003).
Massagué, J. TGFβ in cancer. Cell 134, 215–230 (2008).
Huang, W. et al. Pattern of invasion in human pancreatic cancer organoids is associated with loss of SMAD4 and clinical outcome. Cancer Res. 80, 2804–2817 (2020).
Aung, K. L. et al. Genomics-driven precision medicine for advanced pancreatic cancer: early results from the COMPASS trial. Clin. Cancer Res. 24, 1344–1354 (2018).
O’Kane, G. M. et al. GATA6 expression distinguishes classical and basal-like subtypes in advanced pancreatic cancer. Clin. Cancer Res. 26, 4901–4910 (2020).
Centore, R. C., Sandoval, G. J., Mendes Soares, L. M., Kadoch, C. & Chan, H. M. Mammalian SWI/SNF chromatin remodeling complexes: emerging mechanisms and therapeutic strategies. Trends Genet. 36, 936–950 (2020).
Wang, L. & Shilatifard, A. UTX mutations in human cancer. Cancer Cell 35, 168–176 (2019).
Shain, A. H. et al. Convergent structural alterations define SWItch/Sucrose NonFermentable (SWI/SNF) chromatin remodeler as a central tumor suppressive complex in pancreatic cancer. Proc. Natl Acad. Sci. USA 109, E252–E259 (2012).
Clapier, C. R., Iwasa, J., Cairns, B. R. & Peterson, C. L. Mechanisms of action and regulation of ATP-dependent chromatin-remodelling complexes. Nat. Rev. Mol. Cell Biol. 18, 407–422 (2017).
Rafati, H. et al. Repressive LTR nucleosome positioning by the BAF complex is required for HIV latency. PLoS Biol. 9, e1001206 (2011).
Sze, C. C. & Shilatifard, A. MLL3/MLL4/COMPASS family on epigenetic regulation of enhancer function and cancer. Cold Spring Harb. Perspect. Med. 6, a026427 (2016).
Somerville, T. D. D. et al. TP63-mediated enhancer reprogramming drives the squamous subtype of pancreatic ductal adenocarcinoma. Cell Rep. 25, 1741–1755 (2018).
Somerville, T. D. et al. Squamous trans-differentiation of pancreatic cancer cells promotes stromal inflammation. eLife 9, e53381 (2020).
Roe, J. S. et al. Enhancer reprogramming promotes pancreatic cancer metastasis. Cell 170, 875–888 (2017).
Andricovich, J. et al. Loss of KDM6A activates super-enhancers to induce gender-specific squamous-like pancreatic cancer and confers sensitivity to BET inhibitors. Cancer Cell 33, 512–526 (2018).
Turan, S. & Bastepe, M. GNAS spectrum of disorders. Curr. Osteoporos. Rep. 13, 146–158 (2015).
Dumitrescu, C. E. & Collins, M. T. McCune-Albright syndrome. Orphanet J. Rare Dis. 3, 12 (2008).
Wu, J. et al. Recurrent GNAS mutations define an unexpected pathway for pancreatic cyst development. Sci. Transl Med. 3, 2ra66 (2011).
Hao, H. X., Jiang, X. & Cong, F. Control of Wnt receptor turnover by R-spondin-ZNRF3/RNF43 signaling module and its dysregulation in cancer. Cancers 8, 54 (2016).
Hao, H. X. et al. ZNRF3 promotes Wnt receptor turnover in an R-spondin-sensitive manner. Nature 485, 195–202 (2012).
Springer, S. et al. A combination of molecular markers and clinical features improve the classification of pancreatic cysts. Gastroenterology 149, 1501–1510 (2015).
Bradley, C. A. Guiding pancreatic cyst management. Nat. Rev. Gastroenterol. Hepatol. 16, 582–583 (2019).
Kwei, K. A. et al. Genomic profiling identifies GATA6 as a candidate oncogene amplified in pancreatobiliary cancer. PLoS Genet. 4, e1000081 (2008).
Fu, B., Luo, M., Lakkur, S., Lucito, R. & Iacobuzio-Donahue, C. A. Frequent genomic copy number gain and overexpression of GATA-6 in pancreatic carcinoma. Cancer Biol. Ther. 7, 1593–1601 (2008).
Zhong, Y. et al. GATA6 activates Wnt signaling in pancreatic cancer by negatively regulating the Wnt antagonist Dickkopf-1. PLoS ONE 6, e22129 (2011).
Dang, C. V. MYC on the path to cancer. Cell 149, 22–35 (2012).
Baker, N. E. Emerging mechanisms of cell competition. Nat. Rev. Genet. 21, 683–697 (2020).
Heger, P., Zheng, W., Rottmann, A., Panfilio, K. A. & Wiehe, T. The genetic factors of bilaterian evolution. eLife 9, e45530 (2020).
Ballard, M. S. et al. Mammary stem cell self-renewal is regulated by Slit2/Robo1 signaling through SNAI1 and mINSC. Cell Rep. 13, 290–301 (2015).
Zhang, J. et al. Disease-causing mutations in SF3B1 alter splicing by disrupting interaction with SUGP1. Mol. Cell 76, 82–95 (2019).
Jung, J. H., Lee, H., Zeng, S. X. & Lu, H. RBM10, a new regulator of p53. Cells 9, 2107 (2020).
Matsubayashi, H. et al. Familial pancreatic cancer: concept, management and issues. World J. Gastroenterol. 23, 935–948 (2017).
Klein, A. P. Identifying people at a high risk of developing pancreatic cancer. Nat. Rev. Cancer 3, 66–74 (2013).
Roberts, N. J. et al. ATM mutations in patients with hereditary pancreatic cancer. Cancer Discov. 2, 41–46 (2012).
Roberts, N. J. et al. Whole genome sequencing defines the genetic heterogeneity of familial pancreatic cancer. Cancer Discov. 6, 166–175 (2016).
Shindo, K. et al. Deleterious germline mutations in patients with apparently sporadic pancreatic adenocarcinoma. J. Clin. Oncol. 35, 3382–3390 (2017).
Mizukami, K. et al. Genetic characterization of pancreatic cancer patients and prediction of carrier status of germline pathogenic variants in cancer-predisposing genes. EBioMedicine 60, 103033 (2020).
Lowery, M. A. et al. Prospective evaluation of germline alterations in patients with exocrine pancreatic neoplasms. J. Natl Cancer Inst. 110, 1067–3390 (2018).
Lord, C. J. & Ashworth, A. BRCAness revisited. Nat. Rev. Cancer 16, 110–120 (2016).
Kleeff, J. et al. Chronic pancreatitis. Nat. Rev. Dis. Prim. 3, 17060 (2017).
Shelton, C. A., Umapathy, C., Stello, K., Yadav, D. & Whitcomb, D. C. Hereditary pancreatitis in the United States: survival and rates of pancreatic cancer. Am. J. Gastroenterol. 113, 1376–1384 (2018).
Tamura, K. et al. Mutations in the pancreatic secretory enzymes CPA1 and CPB1 are associated with pancreatic cancer. Proc. Natl Acad. Sci. USA 115, 4767–4772 (2018).
Tomasetti, C., Vogelstein, B. & Parmigiani, G. Half or more of the somatic mutations in cancers of self-renewing tissues originate prior to tumor initiation. Proc. Natl Acad. Sci. USA 110, 1999–2004 (2013).
Yan, L. et al. Molecular analysis to detect pancreatic ductal adenocarcinoma in high-risk groups. Gastroenterology 128, 2124–2130 (2005).
Löhr, M. et al. P53 and K-ras mutations in pancreatic juice samples from patients with chronic pancreatitis. Gastrointest. Endosc. 53, 734–743 (2001).
Martincorena, I. et al. Somatic mutant clones colonize the human esophagus with age. Science 362, 911–917 (2018).
Martincorena, I. et al. High burden and pervasive positive selection of somatic mutations in normal human skin. Science 348, 880–886 (2015).
Guerra, C. et al. Chronic pancreatitis is essential for induction of pancreatic ductal adenocarcinoma by K-Ras oncogenes in adult mice. Cancer Cell 11, 291–302 (2007).
Kandikattu, H. K., Venkateshaiah, S. U. & Mishra, A. Chronic pancreatitis and the development of pancreatic cancer. Endocrine, Metab. Immune Disord. Drug Targets 20, 1182–1210 (2020).
Wang, L. et al. ATDC is required for the initiation of KRAS-induced pancreatic tumorigenesis. Genes Dev. 33, 641–655 (2019).
Ling, J. et al. Kras G12D-induced IKK2/β/NF-κB activation by IL-1α and p62 feedforward loops is required for development of pancreatic ductal adenocarcinoma. Cancer Cell 21, 105–120 (2012).
Kopp, J. L. et al. Identification of Sox9-dependent acinar-to-ductal reprogramming as the principal mechanism for initiation of pancreatic ductal adenocarcinoma. Cancer Cell 22, 737–750 (2012).
Lee, A. Y. L. et al. Cell of origin affects tumour development and phenotype in pancreatic ductal adenocarcinoma. Gut 68, 487–498 (2019).
Shi, C. et al. Differential cell susceptibilities to KrasG12D in the setting of obstructive chronic pancreatitis. Cell. Mol. Gastroenterol. Hepatol. 8, 579–594 (2019).
Pylayeva-Gupta, Y. et al. IL35-producing B cells promote the development of pancreatic neoplasia. Cancer Discov. 6, 247–255 (2016).
Ardito, C. M. et al. EGF receptor is required for KRAS-induced pancreatic tumorigenesis. Cancer Cell 22, 304–317 (2012).
Hermann, P. C. et al. Nicotine promotes initiation and progression of KRAS-induced pancreatic cancer via Gata6-dependent dedifferentiation of acinar cells in mice. Gastroenterology 147, 1119–1133 (2014).
McAllister, F. et al. Oncogenic Kras activates a hematopoietic-to-epithelial IL-17 signaling axis in preinvasive pancreatic neoplasia. Cancer Cell 25, 621–637 (2014).
Luo, Y. et al. Oncogenic KRAS reduces expression of FGF21 in acinar cells to promote pancreatic tumorigenesis in mice on a high-fat diet. Gastroenterology 157, 1413–1428 (2019).
Daniluk, J. et al. An NF-κB pathway-mediated positive feedback loop amplifies Ras activity to pathological levels in mice. J. Clin. Invest. 122, 1519–1528 (2012).
Lahouel, K. et al. Revisiting the tumorigenesis timeline with a data-driven generative model. Proc. Natl Acad. Sci. USA 117, 857–864 (2020).
Matsuda, Y. et al. The prevalence and clinicopathological characteristics of high-grade pancreatic intraepithelial neoplasia autopsy study evaluating the entire pancreatic parenchyma. Pancreas 46, 658–664 (2017).
Wangsa, D. et al. Near-tetraploid cancer cells show chromosome instability triggered by replication stress and exhibit enhanced invasiveness. FASEB J. 32, 3502–3517 (2018).
Van de Peer, Y., Mizrachi, E. & Marchal, K. The evolutionary significance of polyploidy. Nat. Rev. Genet. 18, 411–424 (2017).
Stromnes, I. M., DelGiorno, K. E., Greenberg, P. D. & Hingorani, S. R. Stromal re-engineering to treat pancreas cancer. Carcinogenesis 35, 1451–1460 (2014).
Gurtner, G. C., Werner, S., Barrandon, Y. & Longaker, M. T. Wound repair and regeneration. Nature 35, 314–321 (2008).
Dvorak, H. F. Similarities between tumor stroma generation and wound healing. N. Engl. J. Med. 315, 1650–1659 (1986).
Hutchings, D. et al. Cancerization of the pancreatic ducts: demonstration of a common and under-recognized process using immunolabeling of paired duct lesions and invasive pancreatic ductal adenocarcinoma for p53 and Smad4 expression. Am. J. Surg. Pathol. 42, 1556–1561 (2018).
Makohon-Moore, A. P. et al. Limited heterogeneity of known driver gene mutations among the metastases of individual patients with pancreatic cancer. Nat. Genet. 49, 358–366 (2017).
Reiter, J. G. et al. An analysis of genetic heterogeneity in untreated cancers. Nat. Rev. Cancer 19, 639–650 (2019).
Bechard, M. E. et al. Pancreatic cancers suppress negative feedback of glucose transport to reprogram chromatin for metastasis. Nat. Commun. 11, 4055 (2020).
McDonald, O. G. et al. Epigenomic reprogramming during pancreatic cancer progression links anabolic glucose metabolism to distant metastasis. Nat. Genet. 49, 367–376 (2017).
Pishvaian, M. J. et al. Overall survival in patients with pancreatic cancer receiving matched therapies following molecular profiling: a retrospective analysis of the Know Your Tumor registry trial. Lancet Oncol. 21, 508–518 (2020).
O’Reilly, E. M. et al. Randomized, multicenter, phase II trial of gemcitabine and cisplatin with or without veliparib in patients with pancreas adenocarcinoma and a germline BRCA/PALB2 mutation. J. Clin. Oncol. 38, 1378–1388 (2020).
Golan, T. et al. Maintenance olaparib for germline BRCA-mutated metastatic pancreatic cancer. N. Engl. J. Med. 381, 317–327 (2019).
Hong, D. S. et al. KRAS G12C inhibition with sotorasib in advanced solid tumors. N. Engl. J. Med. 383, 1207–1217 (2020).
Lito, P., Solomon, M., Li, L. S., Hansen, R. & Rosen, N. Cancer therapeutics: allele-specific inhibitors inactivate mutant KRAS G12C by a trapping mechanism. Science 351, 604–608 (2016).
Janes, M. R. et al. Targeting KRAS mutant cancers with a covalent G12C-specific inhibitor. Cell 172, 578–589 (2018).
Chou, A. et al. Clinical and molecular characterization of HER2 amplified-pancreatic cancer. Genome Med. 5, 1–11 (2013).
Wagner, A. D., Özdemir, B. C. & Rüschoff, J. Human epidermal growth factor receptor 2-positive digestive tumors. Curr. Opin. Oncol. 31, 354–361 (2019).
Chaturvedi, S., Hoffman, R. M. & Bertino, J. R. Exploiting methionine restriction for cancer treatment. Biochem. Pharmacol. 154, 170–173 (2018).
Marjon, K. et al. MTAP deletions in cancer create vulnerability to targeting of the MAT2A/PRMT5/RIOK1 axis. Cell Rep. 15, 574–587 (2016).
Basturk, O. et al. A revised classification system and recommendations from the Baltimore Consensus Meeting for neoplastic precursor lesions in the pancreas. Am. J. Surg. Pathol. 39, 1730–1741 (2015).
Yachida, S. & Iacobuzio-Donahue, C. A. Evolution and dynamics of pancreatic cancer progression. Oncogene 32, 5253–5260 (2013).
Hruban, R. H., Goggins, M., Parsons, J. & Kern, S. E. Progression model for pancreatic cancer. Clin. Cancer Res. 6, 2969–2972 (2000).
Matthaei, H., Schulick, R. D., Hruban, R. H. & Maitra, A. Cystic precursors to invasive pancreatic cancer. Nat. Rev. Gastroenterol. Hepatol. 8, 141–150 (2011).
Collisson, E. A., Bailey, P., Chang, D. K. & Biankin, A. V. Molecular subtypes of pancreatic cancer. Nat. Rev. Gastroenterol. Hepatol. 16, 207–220 (2019).
Juiz, N. et al. Basal-like and classical cells coexist in pancreatic cancer revealed by single-cell analysis on biopsy-derived pancreatic cancer organoids from the classical subtype. FASEB J. 34, 12214–12228 (2020).
Miyabayashi, K. et al. Intraductal transplantation models of human pancreatic ductal adenocarcinoma reveal progressive transition of molecular subtypes. Cancer Discov. 10, 1566–1589 (2020).
Nicolle, R. et al. Establishment of a pancreatic adenocarcinoma molecular gradient (PAMG) that predicts the clinical outcome of pancreatic cancer. EBioMedicine 57, 102858 (2020).
Author information
Authors and Affiliations
Contributions
C.A.I.-D. and A.H. contributed to researching data for the article, made a substantial contribution to discussion of content, and wrote and reviewed/edited the manuscript before submission. J.H. wrote and reviewed/edited the manuscript before submission.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information
Nature Reviews Gastroenterology & Hepatology thanks A. Biankin and the other anonymous reviewer(s) for their contribution to the peer review of this work.
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Hayashi, A., Hong, J. & Iacobuzio-Donahue, C.A. The pancreatic cancer genome revisited. Nat Rev Gastroenterol Hepatol 18, 469–481 (2021). https://doi.org/10.1038/s41575-021-00463-z
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41575-021-00463-z
This article is cited by
-
Alteration of chromosome structure impacts gene expressions implicated in pancreatic ductal adenocarcinoma cells
BMC Genomics (2024)
-
An efficient multi-class classification of skin cancer using optimized vision transformer
Medical & Biological Engineering & Computing (2024)
-
Design and evaluation of a multiepitope vaccine for pancreatic cancer using immune-dominant epitopes derived from the signature proteome in expression datasets
Medical Oncology (2024)
-
TP53 missense mutation reveals gain-of-function properties in small-sized KRAS transformed pancreatic ductal adenocarcinoma
Journal of Translational Medicine (2023)
-
Overexpression of ZNF488 supports pancreatic cancer cell proliferation and tumorigenesis through inhibition of ferroptosis via regulating SCD1-mediated unsaturated fatty acid metabolism
Biology Direct (2023)