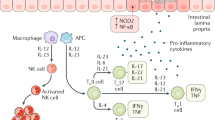
Abstract
Crohn’s disease can affect any part of the gastrointestinal tract; however, current European and national guidelines worldwide do not differentiate between small-intestinal and colonic Crohn’s disease for medical treatment. Data from the past decade provide evidence that ileal Crohn’s disease is distinct from colonic Crohn’s disease in several intestinal layers. Remarkably, colonic Crohn’s disease shows an overlap with regard to disease behaviour with ulcerative colitis, underlining the fact that there is more to inflammatory bowel disease than just Crohn’s disease and ulcerative colitis, and that subtypes, possibly defined by location and shared pathophysiology, are also important. This Review provides a structured overview of the differentiation between ileal and colonic Crohn’s disease using data in the context of epidemiology, genetics, macroscopic differences such as creeping fat and histological findings, as well as differences in regard to the intestinal barrier including gut microbiota, mucus layer, epithelial cells and infiltrating immune cell populations. We also discuss the translation of these basic findings to the clinic, emphasizing the important role of treatment decisions. Thus, this Review provides a conceptual outlook on a new mechanism-driven classification of Crohn’s disease.
Key points
-
Emerging data from fields including clinical behaviour, epidemiology, genetics and the gut microbiota strongly suggest that ileal and colonic Crohn’s disease should be regarded as at least two subtypes of this disease entity.
-
Creeping fat, which represents hyperplasia of the mesenteric fat and wrapping of inflamed intestinal segments, is unique to small-intestinal Crohn’s disease and absent in colonic Crohn’s disease.
-
Predictive models applying a genetic risk score are able to differentiate between ileal and colonic Crohn’s disease.
-
The ileum in Crohn’s disease is characterized by a type 1 or type 17 helper T cell profile, whereas the colon is characterized by a type 1 profile.
-
There is an observable trend that patients with Crohn’s disease with isolated ileal manifestation are less likely to respond to biological therapies than patients with colonic disease.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
References
Torres, J. et al. ECCO guidelines on therapeutics in Crohn’s disease: medical treatment. J. Crohns Colitis 14, 4–22 (2020).
Satsangi, J., Silverberg, M. S., Vermeire, S. & Colombel, J. F. The Montreal classification of inflammatory bowel disease: controversies, consensus, and implications. Gut 55, 749–753 (2006).
Dulai, P. S. et al. Should we divide Crohn’s disease into ileum-dominant and isolated colonic diseases? Clin. Gastroenterol. Hepatol. 17, 2634–2643 (2019).
Subramanian, S., Ekbom, A. & Rhodes, J. M. Recent advances in clinical practice: a systematic review of isolated colonic Crohn’s disease: the third IBD? Gut 66, 362–381 (2017).
Chouraki, V. et al. The changing pattern of Crohn’s disease incidence in northern France: a continuing increase in the 10- to 19-year-old age bracket (1988–2007). Aliment. Pharmacol. Ther. 33, 1133–1142 (2011).
Gunesh, S., Thomas, G. A., Williams, G. T., Roberts, A. & Hawthorne, A. B. The incidence of Crohn’s disease in Cardiff over the last 75 years: an update for 1996-2005. Aliment. Pharmacol. Ther. 27, 211–219 (2008).
Kyle, J. Crohn’s disease in the northeastern and northern Isles of Scotland: an epidemiological review. Gastroenterology 103, 392–399 (1992).
Lapidus, A., Bernell, O., Hellers, G., Persson, P. G. & Lofberg, R. Incidence of Crohn’s disease in Stockholm County 1955-1989. Gut 41, 480–486 (1997).
Cleynen, I. et al. Inherited determinants of Crohn’s disease and ulcerative colitis phenotypes: a genetic association study. Lancet 387, 156–167 (2016). In this study, by testing genotype–phenotype associations, predictive models applying a genetic risk score were able to distinguish ileal Crohn’s disease and colonic Crohn’s disease.
Cleynen, I. et al. Genetic factors conferring an increased susceptibility to develop Crohn’s disease also influence disease phenotype: results from the IBDchip European Project. Gut 62, 1556–1565 (2013).
Cornish, J. A. et al. The risk of oral contraceptives in the etiology of inflammatory bowel disease: a meta-analysis. Am. J. Gastroenterol. 103, 2394–2400 (2008).
Lindberg, E., Jarnerot, G. & Huitfeldt, B. Smoking in Crohn’s disease: effect on localisation and clinical course. Gut 33, 779–782 (1992).
Aldhous, M. C. et al. Does cigarette smoking influence the phenotype of Crohn’s disease? Analysis using the Montreal classification. Am. J. Gastroenterol. 102, 577–588 (2007).
Breuer-Katschinski, B. D., Hollander, N. & Goebell, H. Effect of cigarette smoking on the course of Crohn’s disease. Eur. J. Gastroenterol. Hepatol. 8, 225–228 (1996).
Cosnes, J. et al. Effects of current and former cigarette smoking on the clinical course of Crohn’s disease. Aliment. Pharmacol. Ther. 13, 1403–1411 (1999).
Hancock, L. et al. Clinical and molecular characteristics of isolated colonic Crohn’s disease. Inflamm. Bowel Dis. 14, 1667–1677 (2008).
Holdstock, G., Savage, D., Harman, M. & Wright, R. Should patients with inflammatory bowel disease smoke? Br. Med. J. 288, 362 (1984).
Nunes, T. et al. Smoking does influence disease behaviour and impacts the need for therapy in Crohn’s disease in the biologic era. Aliment. Pharmacol. Ther. 38, 752–760 (2013).
Russel, M. G. et al. Inflammatory bowel disease: is there any relation between smoking status and disease presentation? European Collaborative IBD Study Group. Inflamm. Bowel Dis. 4, 182–186 (1998).
Somerville, K. W., Logan, R. F., Edmond, M. & Langman, M. J. Smoking and Crohn’s disease. Br. Med. J. 289, 954–956 (1984).
Tobin, M. V., Logan, R. F., Langman, M. J., McConnell, R. B. & Gilmore, I. T. Cigarette smoking and inflammatory bowel disease. Gastroenterology 93, 316–321 (1987).
Chivese, T., Esterhuizen, T. M., Basson, A. R. & Watermeyer, G. The influence of second-hand cigarette smoke exposure during childhood and active cigarette smoking on Crohn’s disease phenotype defined by the Montreal classification scheme in a Western Cape population, South Africa. PLoS ONE 10, e0139597 (2015).
Prideaux, L., De Cruz, P., Ng, S. C. & Kamm, M. A. Serological antibodies in inflammatory bowel disease: a systematic review. Inflamm. Bowel Dis. 18, 1340–1355 (2012).
Linskens, R. K. et al. Evaluation of serological markers to differentiate between ulcerative colitis and Crohn’s disease: pANCA, ASCA and agglutinating antibodies to anaerobic coccoid rods. Eur. J. Gastroenterol. Hepatol. 14, 1013–1018 (2002).
Zholudev, A., Zurakowski, D., Young, W., Leichtner, A. & Bousvaros, A. Serologic testing with ANCA, ASCA, and anti-OmpC in children and young adults with Crohn’s disease and ulcerative colitis: diagnostic value and correlation with disease phenotype. Am. J. Gastroenterol. 99, 2235–2241 (2004).
Koutroubakis, I. E. et al. Anti-Saccharomyces cerevisiae mannan antibodies and antineutrophil cytoplasmic autoantibodies in Greek patients with inflammatory bowel disease. Am. J. Gastroenterol. 96, 449–454 (2001).
Quinton, J. F. et al. Anti-Saccharomyces cerevisiae mannan antibodies combined with antineutrophil cytoplasmic autoantibodies in inflammatory bowel disease: prevalence and diagnostic role. Gut 42, 788–791 (1998).
Lakatos, P. L. et al. Pancreatic autoantibodies are associated with reactivity to microbial antibodies, penetrating disease behavior, perianal disease, and extraintestinal manifestations, but not with NOD2/CARD15 or TLR4 genotype in a Hungarian IBD cohort. Inflamm. Bowel Dis. 15, 365–374 (2009).
Bacher, P. et al. Human anti-fungal Th17 immunity and pathology rely on cross-reactivity against Candida albicans. Cell 176, 1340–1355.e15 (2019).
Rodríguez-Sillke, Y. et al. Small intestinal inflammation but not colitis drives pro-inflammators nutritional antigen-specific T-cell response. J. Crohns Colitis 14, S154–S155 (2020).
Jostins, L. et al. Host-microbe interactions have shaped the genetic architecture of inflammatory bowel disease. Nature 491, 119–124 (2012).
Liu, J. Z. et al. Association analyses identify 38 susceptibility loci for inflammatory bowel disease and highlight shared genetic risk across populations. Nat. Genet. 47, 979–986 (2015).
Hampe, J. et al. Association of NOD2 (CARD 15) genotype with clinical course of Crohn’s disease: a cohort study. Lancet 359, 1661–1665 (2002).
Hugot, J. P. et al. Association of NOD2 leucine-rich repeat variants with susceptibility to Crohn’s disease. Nature 411, 599–603 (2001).
Ogura, Y. et al. A frameshift mutation in NOD2 associated with susceptibility to Crohn’s disease. Nature 411, 603–606 (2001).
Ahmad, T. et al. The molecular classification of the clinical manifestations of Crohn’s disease. Gastroenterology 122, 854–866 (2002).
Cuthbert, A. P. et al. The contribution of NOD2 gene mutations to the risk and site of disease in inflammatory bowel disease. Gastroenterology 122, 867–874 (2002).
de la Concha, E. G. et al. Susceptibility to severe ulcerative colitis is associated with polymorphism in the central MHC gene IKBL. Gastroenterology 119, 1491–1495 (2000).
Economou, M., Trikalinos, T. A., Loizou, K. T., Tsianos, E. V. & Ioannidis, J. P. Differential effects of NOD2 variants on Crohn’s disease risk and phenotype in diverse populations: a metaanalysis. Am. J. Gastroenterol. 99, 2393–2404 (2004).
Newman, B. et al. CARD15 and HLA DRB1 alleles influence susceptibility and disease localization in Crohn’s disease. Am. J. Gastroenterol. 99, 306–315 (2004).
Silverberg, M. S. et al. A population- and family-based study of Canadian families reveals association of HLA DRB1*0103 with colonic involvement in inflammatory bowel disease. Inflamm. Bowel Dis. 9, 1–9 (2003).
Caruso, R., Warner, N., Inohara, N. & Nunez, G. NOD1 and NOD2: signaling, host defense, and inflammatory disease. Immunity 41, 898–908 (2014).
Hampe, J. et al. A genome-wide association scan of nonsynonymous SNPs identifies a susceptibility variant for Crohn disease in ATG16L1. Nat. Genet. 39, 207–211 (2007).
Murthy, A. et al. A Crohn’s disease variant in Atg16l1 enhances its degradation by caspase 3. Nature 506, 456–462 (2014).
Cadwell, K. et al. A key role for autophagy and the autophagy gene Atg16l1 in mouse and human intestinal Paneth cells. Nature 456, 259–263 (2008).
Kaser, A. et al. XBP1 links ER stress to intestinal inflammation and confers genetic risk for human inflammatory bowel disease. Cell 134, 743–756 (2008).
Adolph, T. E. et al. Paneth cells as a site of origin for intestinal inflammation. Nature 503, 272–276 (2013).
VanDussen, K. L. et al. Genetic variants synthesize to produce paneth cell phenotypes that define subtypes of Crohn’s disease. Gastroenterology 146, 200–209 (2014).
Crohn, B. B., Ginzburg, L. & Oppenheimer, G. D. Regional ileitis: a pathological and clinical entity. JAMA 99, 1323–1329 (1932).
Kredel, L. I. et al. T-cell composition in ileal and colonic creeping fat – separating ileal from colonic Crohn’s disease. J. Crohns Colitis 13, 79–91 (2019). This study analysed the mesenteric fat of the ileum and colon in Crohn’s disease, revealing adipocyte hyperplasia, fibrosis and a strong immune cell infiltrate as unique to ileal Crohn’s disease. In addition, the ileum is characterized by a TH1:TH17 profile, whereas the colon shows a TH1 profile in the lamina propria.
Mao, R. et al. The mesenteric fat and intestinal muscle interface: creeping fat influencing stricture formation in Crohn’s disease. Inflamm. Bowel Dis. 25, 421–426 (2019).
Batra, A. et al. Mesenteric fat – control site for bacterial translocation in colitis? Mucosal Immunol. 5, 580–591 (2012).
Peyrin-Biroulet, L. et al. Mesenteric fat as a source of C reactive protein and as a target for bacterial translocation in Crohn’s disease. Gut 61, 78–85 (2012).
Siegmund, B. Mesenteric fat in Crohn’s disease: the hot spot of inflammation? Gut 61, 3–5 (2012).
Zulian, A. et al. Visceral adipocytes: old actors in obesity and new protagonists in Crohn’s disease? Gut 61, 86–94 (2012).
Kredel, L. I. et al. Adipokines from local fat cells shape the macrophage compartment of the creeping fat in Crohn’s disease. Gut 62, 852–862 (2013).
Ha, C. W. Y. et al. Translocation of viable gut microbiota to mesenteric adipose drives formation of creeping fat in humans. Cell 183, 666–683.e17 (2020).
Rahier, J. F. et al. Decreased lymphatic vessel density is associated with postoperative endoscopic recurrence in Crohn’s disease. Inflamm. Bowel Dis. 19, 2084–2090 (2013).
Randolph, G. J. et al. Lymphoid aggregates remodel lymphatic collecting vessels that serve mesenteric lymph nodes in Crohn disease. Am. J. Pathol. 186, 3066–3073 (2016).
Randolph, G. J. & Miller, N. E. Lymphatic transport of high-density lipoproteins and chylomicrons. J. Clin. Invest. 124, 929–935 (2014).
Van Kruiningen, H. J. & Colombel, J. F. The forgotten role of lymphangitis in Crohn’s disease. Gut 57, 1–4 (2008).
Thaunat, O., Kerjaschki, D. & Nicoletti, A. Is defective lymphatic drainage a trigger for lymphoid neogenesis? Trends Immunol. 27, 441–445 (2006).
Wehkamp, J. & Stange, E. F. An update review on the paneth cell as key to ileal Crohn’s disease. Front. Immunol. 11, 646 (2020).
Kobayashi, N., Takahashi, D., Takano, S., Kimura, S. & Hase, K. The roles of Peyer’s patches and microfold cells in the gut immune system: relevance to autoimmune diseases. Front. Immunol. 10, 2345 (2019).
Hold, G. L. et al. Role of the gut microbiota in inflammatory bowel disease pathogenesis: what have we learnt in the past 10 years? World J. Gastroenterol. 20, 1192–1210 (2014).
Kostic, A. D., Xavier, R. J. & Gevers, D. The microbiome in inflammatory bowel disease: current status and the future ahead. Gastroenterology 146, 1489–1499 (2014).
Gevers, D. et al. The treatment-naive microbiome in new-onset Crohn’s disease. Cell Host Microbe 15, 382–392 (2014).
Darfeuille-Michaud, A. et al. High prevalence of adherent-invasive Escherichia coli associated with ileal mucosa in Crohn’s disease. Gastroenterology 127, 412–421 (2004). In this study, adherent-invasive Escherichia coli previously identified in the intestinal mucosa of patients with Crohn’s disease was detected in ileal specimens of patients with Crohn’s disease.
Rhodes, J. M. The role of Escherichia coli in inflammatory bowel disease. Gut 56, 610–612 (2007).
Rolhion, N. & Darfeuille-Michaud, A. Adherent-invasive Escherichia coli in inflammatory bowel disease. Inflamm. Bowel Dis. 13, 1277–1283 (2007).
Bringer, M. A., Glasser, A. L., Tung, C. H., Meresse, S. & Darfeuille-Michaud, A. The Crohn’s disease-associated adherent-invasive Escherichia coli strain LF82 replicates in mature phagolysosomes within J774 macrophages. Cell Microbiol. 8, 471–484 (2006).
De la Fuente, M. et al. Escherichia coli isolates from inflammatory bowel diseases patients survive in macrophages and activate NLRP3 inflammasome. Int. J. Med. Microbiol. 304, 384–392 (2014).
Meconi, S. et al. Adherent-invasive Escherichia coli isolated from Crohn’s disease patients induce granulomas in vitro. Cell Microbiol. 9, 1252–1261 (2007).
Barnich, N. et al. CEACAM6 acts as a receptor for adherent-invasive E. coli, supporting ileal mucosa colonization in Crohn disease. J. Clin. Invest. 117, 1566–1574 (2007).
Baumgart, M. et al. Culture independent analysis of ileal mucosa reveals a selective increase in invasive Escherichia coli of novel phylogeny relative to depletion of Clostridiales in Crohn’s disease involving the ileum. ISME J. 1, 403–418 (2007).
Lopez-Siles, M. et al. Mucosa-associated Faecalibacterium prausnitzii and Escherichia coli co-abundance can distinguish irritable bowel syndrome and inflammatory bowel disease phenotypes. Int. J. Med. Microbiol. 304, 464–475 (2014).
Willing, B. et al. Twin studies reveal specific imbalances in the mucosa-associated microbiota of patients with ileal Crohn’s disease. Inflamm. Bowel Dis. 15, 653–660 (2009).
Sokol, H. et al. Faecalibacterium prausnitzii is an anti-inflammatory commensal bacterium identified by gut microbiota analysis of Crohn disease patients. Proc. Natl Acad. Sci. USA 105, 16731–16736 (2008).
Schulthess, J. et al. The short chain fatty acid butyrate imprints an antimicrobial program in macrophages. Immunity 50, 432–445.e7 (2019).
Dicksved, J. et al. Molecular analysis of the gut microbiota of identical twins with Crohn’s disease. ISME J. 2, 716–727 (2008).
Tyler, A. D. et al. Microbiome heterogeneity characterizing intestinal tissue and inflammatory bowel disease phenotype. Inflamm. Bowel Dis. 22, 807–816 (2016).
Willing, B. P. et al. A pyrosequencing study in twins shows that gastrointestinal microbial profiles vary with inflammatory bowel disease phenotypes. Gastroenterology 139, 1844–1854.e1 (2010).
Naftali, T. et al. Distinct microbiotas are associated with ileum-restricted and colon-involving Crohn’s disease. Inflamm. Bowel Dis. 22, 293–302 (2016).
Li, E. et al. Inflammatory bowel diseases phenotype, C. difficile and NOD2 genotype are associated with shifts in human ileum associated microbial composition. PLoS ONE 7, e26284 (2012).
Rajca, S. et al. Alterations in the intestinal microbiome (dysbiosis) as a predictor of relapse after infliximab withdrawal in Crohn’s disease. Inflamm. Bowel Dis. 20, 978–986 (2014).
Kugathasan, S. et al. Prediction of complicated disease course for children newly diagnosed with Crohn’s disease: a multicentre inception cohort study. Lancet 389, 1710–1718 (2017).
Rehman, A. et al. Nod2 is essential for temporal development of intestinal microbial communities. Gut 60, 1354–1362 (2011).
Lavoie, S. et al. The Crohn’s disease polymorphism, ATG16L1 T300A, alters the gut microbiota and enhances the local Th1/Th17 response. eLife 8, e39982 (2019).
Imhann, F. et al. Interplay of host genetics and gut microbiota underlying the onset and clinical presentation of inflammatory bowel disease. Gut 67, 108–119 (2018).
Haberman, Y. et al. Pediatric Crohn disease patients exhibit specific ileal transcriptome and microbiome signature. J. Clin. Invest. 124, 3617–3633 (2014).
Ouellette, A. J. & Bevins, C. L. Paneth cell defensins and innate immunity of the small bowel. Inflamm. Bowel Dis. 7, 43–50 (2001).
Tunzi, C. R. et al. Beta-defensin expression in human mammary gland epithelia. Pediatr. Res. 48, 30–35 (2000).
Johansson, M. E. & Hansson, G. C. Immunological aspects of intestinal mucus and mucins. Nat. Rev. Immunol. 16, 639–649 (2016).
Pelaseyed, T. & Hansson, G. C. Membrane mucins of the intestine at a glance. J. Cell. Sci. 133, jcs240929 (2020).
Allen, A., Hutton, D. A. & Pearson, J. P. The MUC2 gene product: a human intestinal mucin. Int. J. Biochem. Cell Biol. 30, 797–801 (1998).
Axelsson, M. A., Asker, N. & Hansson, G. C. O-Glycosylated MUC2 monomer and dimer from LS 174T cells are water-soluble, whereas larger MUC2 species formed early during biosynthesis are insoluble and contain nonreducible intermolecular bonds. J. Biol. Chem. 273, 18864–18870 (1998).
Pullan, R. D. et al. Thickness of adherent mucus gel on colonic mucosa in humans and its relevance to colitis. Gut 35, 353–359 (1994).
Buisine, M. P. et al. Abnormalities in mucin gene expression in Crohn’s disease. Inflamm. Bowel Dis. 5, 24–32 (1999).
Antoni, L. et al. Human colonic mucus is a reservoir for antimicrobial peptides. J. Crohns Colitis 7, e652–664 (2013).
Bevins, C. L. & Salzman, N. H. Paneth cells, antimicrobial peptides and maintenance of intestinal homeostasis. Nat. Rev. Microbiol. 9, 356–368 (2011).
Ostaff, M. J., Stange, E. F. & Wehkamp, J. Antimicrobial peptides and gut microbiota in homeostasis and pathology. EMBO Mol. Med. 5, 1465–1483 (2013).
Schauber, J. et al. Heterogeneous expression of human cathelicidin hCAP18/LL-37 in inflammatory bowel diseases. Eur. J. Gastroenterol. Hepatol. 18, 615–621 (2006).
Chu, H. et al. Human α-defensin 6 promotes mucosal innate immunity through self-assembled peptide nanonets. Science 337, 477–481 (2012).
Raschig, J. et al. Ubiquitously expressed human beta defensin 1 (hBD1) forms bacteria-entrapping nets in a redox dependent mode of action. PLoS Pathog. 13, e1006261 (2017).
Wehkamp, J. et al. NOD2 (CARD15) mutations in Crohn’s disease are associated with diminished mucosal α-defensin expression. Gut 53, 1658–1664 (2004).
Wehkamp, J. et al. Reduced Paneth cell α-defensins in ileal Crohn’s disease. Proc. Natl Acad. Sci. USA 102, 18129–18134 (2005).
Simms, L. A. et al. Reduced α-defensin expression is associated with inflammation and not NOD2 mutation status in ileal Crohn’s disease. Gut 57, 903–910 (2008).
Liu, T. C. et al. LRRK2 but not ATG16L1 is associated with Paneth cell defect in Japanese Crohn’s disease patients. JCI Insight 2, e91917 (2017).
Tanabe, H. et al. Denatured human α-defensin attenuates the bactericidal activity and the stability against enzymatic digestion. Biochem. Biophys. Res. Commun. 358, 349–355 (2007).
Liu, T. C. et al. Interaction between smoking and ATG16L1T300A triggers Paneth cell defects in Crohn’s disease. J. Clin. Invest. 128, 5110–5122 (2018).
Stappenbeck, T. S. & McGovern, D. P. B. Paneth cell alterations in the development and phenotype of Crohn’s disease. Gastroenterology 152, 322–326 (2017).
Tschurtschenthaler, M. et al. Defective ATG16L1-mediated removal of IRE1α drives Crohn’s disease-like ileitis. J. Exp. Med. 214, 401–422 (2017).
Courth, L. F. et al. Crohn’s disease-derived monocytes fail to induce Paneth cell defensins. Proc. Natl Acad. Sci. USA 112, 14000–14005 (2015).
Peyrin-Biroulet, L. et al. Peroxisome proliferator-activated receptor gamma activation is required for maintenance of innate antimicrobial immunity in the colon. Proc. Natl Acad. Sci. USA 107, 8772–8777 (2010).
Wehkamp, J. et al. Inducible and constitutive β-defensins are differentially expressed in Crohn’s disease and ulcerative colitis. Inflamm. Bowel Dis. 9, 215–223 (2003).
Lala, S. et al. Crohn’s disease and the NOD2 gene: a role for paneth cells. Gastroenterology 125, 47–57 (2003).
Ogura, Y. et al. Expression of NOD2 in Paneth cells: a possible link to Crohn’s ileitis. Gut 52, 1591–1597 (2003).
Wang, Y. et al. Single-cell transcriptome analysis reveals differential nutrient absorption functions in human intestine. J. Exp. Med. 217, e20191130 (2020).
Gunther, C. et al. Interferon lambda promotes paneth cell death via STAT1 signaling in mice and is increased in inflamed ileal tissues of patients with Crohn’s disease. Gastroenterology 157, 1310–1322.e13 (2019). This study shows an increase in IFNλ in serum and inflamed ileum of patients with Crohn’s disease that is associated with a loss of Paneth cells. In mice, secreted IFNλ resulted in a loss of Paneth cells via STAT1 and MLKL in a caspase 8-dependent manner.
Howell, K. J. et al. DNA methylation and transcription patterns in intestinal epithelial cells from pediatric patients with inflammatory bowel diseases differentiate disease subtypes and associate with outcome. Gastroenterology 154, 585–598 (2018). IECs were isolated from mucosal biopsies (ileum, ascending colon, sigmoid) of children with newly diagnosed IBD; changes in DNA methylation and transcription pattern were exclusively identified in IECs from the terminal ileum.
Pierre, N. et al. Proteomics highlights common and distinct pathophysiological processes associated with ileal and colonic ulcers in Crohn’s disease. J. Crohns Colitis 14, 205–215 (2020).
Duvoisin, G. et al. Novel biomarkers and the future potential of biomarkers in inflammatory bowel disease. Mediators Inflamm. 2017, 1936315 (2017).
Kofla-Dlubacz, A., Matusiewicz, M., Krzystek-Korpacka, M. & Iwanczak, B. Correlation of MMP-3 and MMP-9 with Crohn’s disease activity in children. Dig. Dis. Sci. 57, 706–712 (2012).
Verdier, J., Begue, B., Cerf-Bensussan, N. & Ruemmele, F. M. Compartmentalized expression of Th1 and Th17 cytokines in pediatric inflammatory bowel diseases. Inflamm. Bowel Dis. 18, 1260–1266 (2012).
Becker, C. et al. Constitutive p40 promoter activation and IL-23 production in the terminal ileum mediated by dendritic cells. J. Clin. Invest. 112, 693–706 (2003).
Ivanov, I. I. et al. Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell 139, 485–498 (2009).
Monteleone, G. et al. Control of matrix metalloproteinase production in human intestinal fibroblasts by interleukin 21. Gut 55, 1774–1780 (2006).
Monteleone, G. et al. Interleukin-21 enhances T-helper cell type I signaling and interferon-γ production in Crohn’s disease. Gastroenterology 128, 687–694 (2005).
Neurath, M. F. Cytokines in inflammatory bowel disease. Nat. Rev. Immunol. 14, 329–342 (2014).
Pickert, G. et al. STAT3 links IL-22 signaling in intestinal epithelial cells to mucosal wound healing. J. Exp. Med. 206, 1465–1472 (2009).
Leung, J. M. et al. IL-22-producing CD4+ cells are depleted in actively inflamed colitis tissue. Mucosal Immunol. 7, 124–133 (2014).
Ouyang, W., Kolls, J. K. & Zheng, Y. The biological functions of T helper 17 cell effector cytokines in inflammation. Immunity 28, 454–467 (2008).
Sonnenberg, G. F., Fouser, L. A. & Artis, D. Border patrol: regulation of immunity, inflammation and tissue homeostasis at barrier surfaces by IL-22. Nat. Immunol. 12, 383–390 (2011).
Brand, S. et al. IL-22 is increased in active Crohn’s disease and promotes proinflammatory gene expression and intestinal epithelial cell migration. Am. J. Physiol. Gastrointest. Liver Physiol. 290, G827–G838 (2006).
Allez, M. et al. T cell clonal expansions in ileal Crohn’s disease are associated with smoking behaviour and postoperative recurrence. Gut 68, 1961–1970 (2019). A T cell receptor (TCR) analysis was performed of the ileum of patients with Crohn’s disease before and after ileocaecal resection, identifying a subgroup with a persistence of similar TCR repertoire, which was associated with smoking and recurrent disease.
Cosnes, J. et al. Long-term evolution of disease behavior of Crohn’s disease. Inflamm. Bowel Dis. 8, 244–250 (2002).
Feagan, B. G. et al. Ustekinumab as induction and maintenance therapy for Crohn’s disease. N. Engl. J. Med. 375, 1946–1960 (2016).
Sandborn, W. J. et al. Vedolizumab as induction and maintenance therapy for Crohn’s disease. N. Engl. J. Med. 369, 711–721 (2013).
Sandborn, W. J. et al. Adalimumab for maintenance treatment of Crohn’s disease: results of the CLASSIC II trial. Gut 56, 1232–1239 (2007).
Schmitt, H. et al. Expansion of IL-23 receptor bearing TNFR2+ T cells is associated with molecular resistance to anti-TNF therapy in Crohn’s disease. Gut 68, 814–828 (2019).
Visekruna, A. et al. Intestinal development and homeostasis require activation and apoptosis of diet-reactive T cells. J. Clin. Invest. 129, 1972–1983 (2019). In this study, a decreased frequency of apoptotic CD4+ T cells was observed in Peyer’s patches of patients with Crohn’s disease, revealing a potential disease mechanism unique to the ileum.
Habtezion, A., Nguyen, L. P., Hadeiba, H. & Butcher, E. C. Leukocyte trafficking to the small intestine and colon. Gastroenterology 150, 340–354 (2016).
Hynes, R. O. Integrins: bidirectional, allosteric signaling machines. Cell 110, 673–687 (2002).
Shattil, S. J., Kim, C. & Ginsberg, M. H. The final steps of integrin activation: the end game. Nat. Rev. Mol. Cell Biol. 11, 288–300 (2010).
Zundler, S., Becker, E., Schulze, L. L. & Neurath, M. F. Immune cell trafficking and retention in inflammatory bowel disease: mechanistic insights and therapeutic advances. Gut 68, 1688–1700 (2019).
Berlin, C. et al. α4β7 integrin mediates lymphocyte binding to the mucosal vascular addressin MAdCAM-1. Cell 74, 185–195 (1993).
Elices, M. J. et al. VCAM-1 on activated endothelium interacts with the leukocyte integrin VLA-4 at a site distinct from the VLA-4/fibronectin binding site. Cell 60, 577–584 (1990).
Taniguchi, T. et al. Effects of the anti-ICAM-1 monoclonal antibody on dextran sodium sulphate-induced colitis in rats. J. Gastroenterol. Hepatol. 13, 945–949 (1998).
Salmi, M. et al. Immune cell trafficking in uterus and early life is dominated by the mucosal addressin MAdCAM-1 in humans. Gastroenterology 121, 853–864 (2001).
Ley, K., Laudanna, C., Cybulsky, M. I. & Nourshargh, S. Getting to the site of inflammation: the leukocyte adhesion cascade updated. Nat. Rev. Immunol. 7, 678–689 (2007).
Kinashi, T. Intracellular signalling controlling integrin activation in lymphocytes. Nat. Rev. Immunol. 5, 546–559 (2005).
Denucci, C. C., Mitchell, J. S. & Shimizu, Y. Integrin function in T-cell homing to lymphoid and nonlymphoid sites: getting there and staying there. Crit. Rev. Immunol. 29, 87–109 (2009).
Svensson, M. et al. CCL25 mediates the localization of recently activated CD8αβ+ lymphocytes to the small-intestinal mucosa. J. Clin. Invest. 110, 1113–1121 (2002).
Fischer, A. et al. Differential effects of α4β7 and GPR15 on homing of effector and regulatory T cells from patients with UC to the inflamed. gut in vivo. Gut 65, 1642–1664 (2016).
Kim, S. V. et al. GPR15-mediated homing controls immune homeostasis in the large intestine mucosa. Science 340, 1456–1459 (2013). This study identified GPR15, an orphan heterotrimeric guanine nucleotide-binding protein (G-protein)-coupled receptor as the homing receptor in particular for T cells to the lamina propria of the colon.
Nguyen, L. P. et al. Role and species-specific expression of colon T cell homing receptor GPR15 in colitis. Nat. Immunol. 16, 207–213 (2015).
Ocon, B. et al. A mucosal and cutaneous chemokine ligand for the lymphocyte chemoattractant receptor GPR15. Front. Immunol. 8, 1111 (2017).
Suply, T. et al. A natural ligand for the orphan receptor GPR15 modulates lymphocyte recruitment to epithelia. Sci. Signal 10, eaal0180 (2017).
Zundler, S. et al. Blockade of αEβ7 integrin suppresses accumulation of CD8(+) and Th9 lymphocytes from patients with IBD in the inflamed gut in vivo. Gut 66, 1936–1948 (2017).
Kotze, P. G. et al. Real-world clinical, endoscopic and radiographic efficacy of vedolizumab for the treatment of inflammatory bowel disease. Aliment. Pharmacol. Ther. 48, 626–637 (2018).
Louis, E. et al. Early development of stricturing or penetrating pattern in Crohn’s disease is influenced by disease location, number of flares, and smoking but not by NOD2/CARD15 genotype. Gut 52, 552–557 (2003).
Thia, K. T., Sandborn, W. J., Harmsen, W. S., Zinsmeister, A. R. & Loftus, E. V. Jr. Risk factors associated with progression to intestinal complications of Crohn’s disease in a population-based cohort. Gastroenterology 139, 1147–1155 (2010).
Guizzetti, L. et al. Development of clinical prediction models for surgery and complications in Crohn’s disease. J. Crohns Colitis 12, 167–177 (2018).
Ott, C. & Scholmerich, J. Extraintestinal manifestations and complications in IBD. Nat. Rev. Gastroenterol. Hepatol. 10, 585–595 (2013).
Vavricka, S. R. et al. Extraintestinal manifestations of inflammatory bowel disease. Inflamm. Bowel Dis. 21, 1982–1992 (2015).
Ardizzone, S., Puttini, P. S., Cassinotti, A. & Porro, G. B. Extraintestinal manifestations of inflammatory bowel disease. Dig. Liver Dis. 40 (Suppl. 2), S253–S259 (2008).
Farhi, D. et al. Significance of erythema nodosum and pyoderma gangrenosum in inflammatory bowel diseases: a cohort study of 2402 patients. Medicine 87, 281–293 (2008).
Bhagat, S. & Das, K. M. A shared and unique peptide in the human colon, eye, and joint detected by a monoclonal antibody. Gastroenterology 107, 103–108 (1994).
Boonstra, K. et al. Primary sclerosing cholangitis is associated with a distinct phenotype of inflammatory bowel disease. Inflamm. Bowel Dis. 18, 2270–2276 (2012).
Iny, O. et al. Crohn’s disease behavior and location is altered when associated with primary sclerosing cholangitis. Isr. Med. Assoc. J. 20, 25–29 (2018).
Hanauer, S. B. et al. Maintenance infliximab for Crohn’s disease: the ACCENT I randomised trial. Lancet 359, 1541–1549 (2002).
Targan, S. R. et al. A short-term study of chimeric monoclonal antibody cA2 to tumor necrosis factor α for Crohn’s disease. N. Engl. J. Med. 337, 1029–1036 (1997).
Hanauer, S. B. et al. Human anti-tumor necrosis factor monoclonal antibody (adalimumab) in Crohn’s disease: the CLASSIC-I trial. Gastroenterology 130, 323–333 (2006).
Sandborn, W. J. et al. Certolizumab pegol for active Crohn’s disease: a placebo-controlled, randomized trial. Clin. Gastroenterol. Hepatol. 9, 670–678.e3 (2011).
Arnott, I. D., McNeill, G. & Satsangi, J. An analysis of factors influencing short-term and sustained response to infliximab treatment for Crohn’s disease. Aliment. Pharmacol. Ther. 17, 1451–1457 (2003).
Laharie, D. et al. Predictors of response to infliximab in luminal Crohn’s disease. Gastroenterol. Clin. Biol. 29, 145–149 (2005).
Vermeire, S. et al. Demographic and clinical parameters influencing the short-term outcome of anti-tumor necrosis factor (infliximab) treatment in Crohn’s disease. Am. J. Gastroenterol. 97, 2357–2363 (2002).
Reinisch, W. et al. Characterisation of mucosal healing with adalimumab treatment in patients with moderately to severely active Crohn’s disease: results from the EXTEND trial. J. Crohns Colitis 11, 425–434 (2017).
Takenaka, K. et al. Small bowel healing detected by endoscopy in patients with Crohn’s disease after treatment with antibodies against tumor necrosis factor. Clin. Gastroenterol. Hepatol. 18, 1545–1552 (2020).
Danese, S. et al. Endoscopic, radiologic, and histologic healing with vedolizumab in patients with active Crohn’s disease. Gastroenterology 157, 1007–1018 e1007 (2019). This prospective, open-label, phase 3b study evaluated endoscopic, radiological and histological healing in the ileum and colon upon vedolizumab treatment in patients with Crohn’s disease.
Feagan, B. G. et al. Induction therapy with the selective interleukin-23 inhibitor risankizumab in patients with moderate-to-severe Crohn’s disease: a randomised, double-blind, placebo-controlled phase 2 study. Lancet 389, 1699–1709 (2017).
Visvanathan, S. et al. Selective IL-23 inhibition by risankizumab modulates the molecular profile in the colon and ileum of patients with active Crohn’s disease: results from a randomised phase II biopsy sub-study. J. Crohns Colitis 12, 1170–1179 (2018).
Atreya, R. & Neurath, M. F. Mechanisms of molecular resistance and predictors of response to biological therapy in inflammatory bowel disease. Lancet Gastroenterol. Hepatol. 3, 790–802 (2018).
Atreya, R., Neurath, M. F. & Siegmund, B. Personalizing treatment in IBD: hype or reality in 2020? Can we predict response to anti-TNF? Front. Med. 7, 517 (2020).
Gisbert, J. P. & Chaparro, M. Predictors of primary response to biologic treatment [anti-TNF, vedolizumab, and ustekinumab] in patients with inflammatory bowel disease: from basic science to clinical practice. J. Crohns Colitis 14, 694–709 (2020).
Arijs, I. et al. Predictive value of epithelial gene expression profiles for response to infliximab in Crohn’s disease. Inflamm. Bowel Dis. 16, 2090–2098 (2010).
Arijs, I. et al. Mucosal gene signatures to predict response to infliximab in patients with ulcerative colitis. Gut 58, 1612–1619 (2009).
Brubaker, D. K. et al. An interspecies translation model implicates integrin signaling in infliximab-resistant inflammatory bowel disease. Sci. Signal 13, eaay3258 (2020).
Belarif, L. et al. IL-7 receptor influences anti-TNF responsiveness and T cell gut homing in inflammatory bowel disease. J. Clin. Invest. 129, 1910–1925 (2019).
Zundler, S. et al. The α4β1 homing pathway is essential for ileal homing of Crohn’s disease effector T cells in vivo. Inflamm. Bowel Dis. 23, 379–391 (2017). This study indicates that homing of effector T cells to the ileum through the α4β1–VCAM1 axis presents an essential and non-redundant pathway for Crohn’s disease in vivo.
Tew, G. W. et al. Association between response to etrolizumab and expression of integrin αE and granzyme A in colon biopsies of patients with ulcerative colitis. Gastroenterology 150, 477–487.e9 (2016).
Ichikawa, R. et al. AlphaE integrin expression is increased in the ileum relative to the colon and unaffected by inflammation. J. Crohns Colitis 12, 1191–1199 (2018). This study demonstrates markedly higher numbers of ileal compared to colonic αE+ cells, which might serve as a predictive biomarker for effectiveness of etrolizumab therapy.
Acknowledgements
R.A. and B.S. are supported by the research initiatives CRC-TRR 241, SI 749/10-1 and SPP1656 (B.S.), CRC1181 (R.A.), and CRC1340 and CRC1449 (B.S.) of the German Research Foundation (DFG). The DFG funds the Heisenberg Professorship of R.A.
Author information
Authors and Affiliations
Contributions
The authors contributed equally to all aspects of this article.
Corresponding author
Ethics declarations
Competing interests
B.S. has served as a consultant for Abbvie, Boehringer, Celgene, Falk, Janssen, Lilly, Pfizer, Prometheus, Takeda and has received speaker’s fees from Abbvie, CED Service GmbH, Falk, Ferring, Janssen, Novartis and Takeda as a representative of Charité – Universitätsmedizin Berlin. R.A. has served as a speaker, or consultant, or received research grants from AbbVie, Biogen, Boehringer Ingelheim, Celgene, Dr. Falk Pharma, Ferring, InDex Pharmaceuticals, Janssen-Cilag, MSD Sharp & Dome, Pfizer, Roche Pharma, Samsung Bioepis and Takeda.
Additional information
Peer review information
Nature Reviews Gastroenterology & Hepatology thanks the anonymous reviewers for their contribution to the peer review of this work.
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Atreya, R., Siegmund, B. Location is important: differentiation between ileal and colonic Crohn’s disease. Nat Rev Gastroenterol Hepatol 18, 544–558 (2021). https://doi.org/10.1038/s41575-021-00424-6
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41575-021-00424-6
This article is cited by
-
Efficacy and safety of gut microbiota-based therapies in autoimmune and rheumatic diseases: a systematic review and meta-analysis of 80 randomized controlled trials
BMC Medicine (2024)
-
Radiological biomarkers reflecting visceral fat distribution help distinguish inflammatory bowel disease subtypes: a multicenter cross-sectional study
Insights into Imaging (2024)
-
Transcriptome-wide association studies associated with Crohn’s disease: challenges and perspectives
Cell & Bioscience (2024)
-
Mitochondrial DNA copy number is associated with Crohn’s disease: a comprehensive Mendelian randomization analysis
Scientific Reports (2023)
-
Prädiktion von Krankheitsverlauf und Therapieerfolg
Die Gastroenterologie (2023)