Abstract
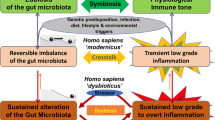
The gut microbiota is a dense and diverse ecosystem that is involved in many physiological functions as well as in disease pathogenesis. It is dominated by bacteria, which have been extensively studied in the past 15 years; however, other microorganisms, such as fungi, phages, archaea and protists, are also present in the gut microbiota. Exploration of the fungal component, namely, the mycobiota, is at an early stage, and several specific technical challenges are associated with mycobiota analysis. The number of fungi in the lower gastrointestinal tract is far lower than that of bacteria, but fungal cells are much larger and much more complex than bacterial cells. In addition, a role of the mycobiota in disease, notably in IBD, is indicated by both descriptive data in humans and mechanistic data in mice. Interactions between bacteria and fungi within the gut, their functional roles and their interplay with the host and its immune system are fascinating areas that researchers are just beginning to investigate. In this Review, we discuss the newest data on the gut mycobiota and explore both the technical aspects of its study and its role in health and gastrointestinal diseases.
Key points
-
Interest in the study of the fungal microbiota has been rising over the past decade, resulting in the accumulation of various data sets that describe the mycobiota in health and disease.
-
This young research area requires standardization of techniques and bioinformatic analysis, as well as complete, curated databases, to reach a level of insight similar to that of the bacterial microbiota.
-
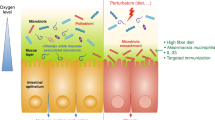
Similar to the bacterial microbiota, environmental conditions, in particular diet, considerably influence the fungal microbiota.
-
Deciphering the multitude of interactions between bacteria and fungi in the gut and in other niches is one of the most promising areas of investigation for gut microbiota manipulation.
-
Many findings now demonstrate that the gut mycobiota can strongly influence the host immune system, but considerable research is still needed to better characterize these interactions.
-
Current observations of the direct or indirect effects of the fungal microbiota on gastrointestinal diseases advocate further investigation of the mycobiota composition and means of controlling its diversity.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Sender, R., Fuchs, S. & Milo, R. Revised estimates for the number of human and bacteria cells in the body. PLOS Biol. 14, e1002533 (2016).
Richard, M. L., Lamas, B., Liguori, G., Hoffmann, T. W. & Sokol, H. Gut fungal microbiota: the Yin and Yang of inflammatory bowel disease. Inflamm. Bowel Dis. 21, 656–665 (2015).
Scupham, A. J. et al. Abundant and diverse fungal microbiota in the murine intestine. Appl. Environ. Microbiol. 72, 793–801 (2006).
Ott, S. J. et al. Fungi and inflammatory bowel diseases: alterations of composition and diversity. Scand. J. Gastroenterol. 43, 831–841 (2008).
Scanlan, P. D. & Marchesi, J. R. Micro-eukaryotic diversity of the human distal gut microbiota: qualitative assessment using culture-dependent and -independent analysis of faeces. ISME J. 2, 1183–1193 (2008).
Ghannoum, M. A. et al. Characterization of the oral fungal microbiome (mycobiome) in healthy individuals. PLOS Pathog. 6, e1000713 (2010).
Mukherjee, P. K. et al. Oral mycobiome analysis of HIV-infected patients: identification of Pichia as an antagonist of opportunistic fungi. PLOS Pathog. 10, e1003996 (2014).
Ackerman, A. L. & Underhill, D. M. The mycobiome of the human urinary tract: potential roles for fungi in urology. Ann. Transl Med. 5, 31 (2017).
Bradford, L. L. & Ravel, J. The vaginal mycobiome: a contemporary perspective on fungi in women’s health and diseases. Virulence 8, 342–351 (2017).
Tipton, L., Ghedin, E. & Morris, A. The lung mycobiome in the next-generation sequencing era. Virulence 8, 334–341 (2017).
Ward, T. L., Knights, D. & Gale, C. A. Infant fungal communities: current knowledge and research opportunities. BMC Med. 15, 30 (2017).
Wheeler, M. L., Limon, J. J. & Underhill, D. M. Immunity to commensal fungi: detente and disease. Annu. Rev. Pathol. 12, 359–385 (2017).
Huseyin, C. E., O’Toole, P. W., Cotter, P. D. & Scanlan, P. D. Forgotten fungi-the gut mycobiome in human health and disease. FEMS Microbiol. Rev. 41, 479–511 (2017).
Mukherjee, P. K. et al. Mycobiota in gastrointestinal diseases. Nat. Rev. Gastroenterol. Hepatol. 12, 77–87 (2015).
Borges, F. M. et al. Fungal diversity of human gut microbiota among eutrophic, overweight, and obese individuals based on aerobic culture-dependent approach. Curr. Microbiol. 75, 726–735 (2018).
Hamad, I. et al. Culturomics and amplicon-based metagenomic approaches for the study of fungal population in human gut microbiota. Sci. Rep. 7, 16788 (2017).
Gouba, N., Raoult, D. & Drancourt, M. Plant and fungal diversity in gut microbiota as revealed by molecular and culture investigations. PLOS ONE 8, e59474 (2013).
Becker, P. T. et al. Identification of filamentous fungi isolates by MALDI-TOF mass spectrometry: clinical evaluation of an extended reference spectra library. Med. Mycol. 52, 826–834 (2014).
Sanguinetti, M. & Posteraro, B. Identification of molds by matrix-assisted laser desorption ionization-time of flight mass spectrometry. J. Clin. Microbiol. 55, 369–379 (2017).
Qin, J. et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature 464, 59–65 (2010).
Donovan, P. D., Gonzalez, G., Higgins, D. G., Butler, G. & Ito, K. Identification of fungi in shotgun metagenomics datasets. PLOS ONE 13, e0192898 (2018).
Lewis, J. D. et al. Inflammation, antibiotics, and diet as environmental stressors of the gut microbiome in pediatric Crohn’s disease. Cell Host Microbe 18, 489–500 (2015).
Vesty, A., Biswas, K., Taylor, M. W., Gear, K. & Douglas, R. G. Evaluating the impact of DNA extraction method on the representation of human oral bacterial and fungal communities. PLOS ONE 12, e0169877 (2017).
Huseyin, C. E., Rubio, R. C., O’Sullivan, O., Cotter, P. D. & Scanlan, P. D. The fungal frontier: a comparative analysis of methods used in the study of the human gut mycobiome. Front. Microbiol. 8, 1432 (2017).
Costea, P. I. et al. Towards standards for human fecal sample processing in metagenomic studies. Nat. Biotechnol. 35, 1069–1076 (2017).
Schoch, C. L. et al. Nuclear ribosomal internal transcribed spacer (ITS) region as a universal DNA barcode marker for Fungi. Proc. Natl Acad. Sci. USA 109, 6241–6246 (2012).
Bellemain, E. et al. ITS as an environmental DNA barcode for fungi: an in silico approach reveals potential PCR biases. BMC Microbiol. 10, 189 (2010).
De Filippis, F., Laiola, M., Blaiotta, G. & Ercolini, D. Different amplicon targets for sequencing-based studies of fungal diversity. Appl. Environ. Microbiol. 83, e00905–17 (2017).
Motooka, D. et al. Fungal ITS1 deep-sequencing strategies to reconstruct the composition of a 26-species community and evaluation of the gut mycobiota of healthy Japanese individuals. Front. Microbiol. 8, 238 (2017).
Usyk, M., Zolnik, C. P., Patel, H., Levi, M. H. & Burk, R. D. Novel ITS1 fungal primers for characterization of the mycobiome. mSphere 2, e00488–17 (2017).
Herrera, M. L., Vallor, A. C., Gelfond, J. A., Patterson, T. F. & Wickes, B. L. Strain-dependent variation in 18S ribosomal DNA copy numbers in Aspergillus fumigatus. J. Clin. Microbiol. 47, 1325–1332 (2009).
Kembel, S. W., Wu, M., Eisen, J. A. & Green, J. L. Incorporating 16S gene copy number information improves estimates of microbial diversity and abundance. PLOS Comput. Biol. 8, e1002743 (2012).
Stielow, J. B. et al. One fungus, which genes? Development and assessment of universal primers for potential secondary fungal DNA barcodes. Persoonia 35, 242–263 (2015).
Tang, J., Iliev, I. D., Brown, J., Underhill, D. M. & Funari, V. A. Mycobiome: approaches to analysis of intestinal fungi. J. Immunol. Methods 421, 112–121 (2015).
Kumar, S. et al. CLOTU: an online pipeline for processing and clustering of 454 amplicon reads into OTUs followed by taxonomic annotation. BMC Bioinformatics 12, 182 (2011).
White, J. R., Maddox, C., White, O., Angiuoli, S. V. & Fricke, W. F. CloVR-ITS: Automated internal transcribed spacer amplicon sequence analysis pipeline for the characterization of fungal microbiota. Microbiome 1, 6 (2013).
Caporaso, J. G. et al. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 7, 335–336 (2010).
Edgar, R. C. Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26, 2460–2461 (2010).
Schloss, P. D. et al. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 75, 7537–7541 (2009).
Gdanetz, K., Benucci, G. M. N., Vande Pol, N. & Bonito, G. CONSTAX: a tool for improved taxonomic resolution of environmental fungal ITS sequences. BMC Bioinformatics 18, 538 (2017).
Palmer, J. M., Jusino, M. A., Banik, M. T. & Lindner, D. L. Non-biological synthetic spike-in controls and the AMPtk software pipeline improve mycobiome data. PeerJ 6, e4925 (2018).
Arbefeville, S., Harris, A. & Ferrieri, P. Comparison of sequencing the D2 region of the large subunit ribosomal RNA gene (MicroSEQ(R)) versus the internal transcribed spacer (ITS) regions using two public databases for identification of common and uncommon clinically relevant fungal species. J. Microbiol. Methods 140, 40–46 (2017).
Nilsson, R. H. et al. Taxonomic reliability of DNA sequences in public sequence databases: a fungal perspective. PLOS ONE 1, e59 (2006).
Irinyi, L. et al. International Society of Human and Animal Mycology (ISHAM)-ITS reference DNA barcoding database—the quality controlled standard tool for routine identification of human and animal pathogenic fungi. Med. Mycol. 53, 313–337 (2015).
Koljalg, U. et al. Towards a unified paradigm for sequence-based identification of fungi. Mol. Ecol. 22, 5271–5277 (2013).
Ratnasingham, S. & Hebert, P. D. N. bold: the barcode of life data system (http://www.barcodinglife.org). Mol. Ecol. Notes 7, 355–364 (2007).
Schoch, C. L. et al. Finding needles in haystacks: linking scientific names, reference specimens and molecular data for fungi. Database 2014, bau061 (2014).
Nilsson, R. H. et al. Taxonomic annotation of public fungal ITS sequences from the built environment — a report from an April 10–11, 2017 workshop (Aberdeen, UK). Mycokeys 28, 65–82 (2018).
Prakash, P. Y. et al. Online databases for taxonomy and identification of pathogenic fungi and proposal for a cloud-based dynamic data network platform. J. Clin. Microbiol. 55, 1011–1024 (2017).
Sekirov, I., Russell, S. L., Antunes, L. C. & Finlay, B. B. Gut microbiota in health and disease. Physiol. Rev. 90, 859–904 (2010).
Jiang, T. T. et al. Commensal fungi recapitulate the protective benefits of intestinal bacteria. Cell Host Microbe 22, 809–816 (2017).
Donaldson, G. P., Lee, S. M. & Mazmanian, S. K. Gut biogeography of the bacterial microbiota. Nat. Rev. Microbiol. 14, 20–32 (2016).
Hallen-Adams, H. E., Kachman, S. D., Kim, J., Legge, R. M. & Martinez, I. Fungi inhabiting the healthy human gastrointestinal tract: a diverse and dynamic community. Fungal Ecol. 15, 9–17 (2015).
Nash, A. K. et al. The gut mycobiome of the Human Microbiome Project healthy cohort. Microbiome 5, 153 (2017).
Hallen-Adams, H. E. & Suhr, M. J. Fungi in the healthy human gastrointestinal tract. Virulence 8, 352–358 (2017).
Suhr, M. J. & Hallen-Adams, H. E. The human gut mycobiome: pitfalls and potentials—a mycologist’s perspective. Mycologia 107, 1057–1073 (2015).
Auchtung, T. A. et al. Investigating colonization of the healthy adult gastrointestinal tract by fungi. mSphere 3, e00092–18 (2018).
Strati, F. et al. Age and gender affect the composition of fungal population of the human gastrointestinal tract. Front. Microbiol. 7, 1227 (2016).
LaTuga, M. S. et al. Beyond bacteria: a study of the enteric microbial consortium in extremely low birth weight infants. PLOS ONE 6, e27858 (2011).
Heisel, T. et al. Complementary amplicon-based genomic approaches for the study of fungal communities in humans. PLOS ONE 10, e0116705 (2015).
Schei, K. et al. Early gut mycobiota and mother-offspring transfer. Microbiome 5, 107 (2017).
Bliss, J. M., Basavegowda, K. P., Watson, W. J., Sheikh, A. U. & Ryan, R. M. Vertical and horizontal transmission of Candida albicans in very low birth weight infants using DNA fingerprinting techniques. Pediatr. Infect. Dis. J. 27, 231–235 (2008).
Nagata, R. et al. Transmission of the major skin microbiota, Malassezia, from mother to neonate. Pediatr. Int. 54, 350–355 (2012).
Ward, T. L. et al. Development of the human mycobiome over the first month of life and across body sites. mSystems 3, e00140–17 (2018).
Boix-Amoros, A., Martinez-Costa, C., Querol, A., Collado, M. C. & Mira, A. Multiple approaches detect the presence of fungi in human breastmilk samples from healthy mothers. Sci. Rep. 7, 13016 (2017).
David, L. A. et al. Diet rapidly and reproducibly alters the human gut microbiome. Nature 505, 559–563 (2014).
Hoffmann, C. et al. Archaea and fungi of the human gut microbiome: correlations with diet and bacterial residents. PLOS ONE 8, e66019 (2013).
Suhr, M. J., Banjara, N. & Hallen-Adams, H. E. Sequence-based methods for detecting and evaluating the human gut mycobiome. Lett. Appl. Microbiol. 62, 209–215 (2016).
Heisel, T. et al. High-fat diet changes fungal microbiomes and interkingdom relationships in the murine gut. mSphere 2, e00351–17 (2017).
Bamford, C. V. et al. Streptococcus gordonii modulates Candida albicans biofilm formation through intergeneric communication. Infect. Immun. 77, 3696–3704 (2009).
Hwang, G. et al. Candida albicans mannans mediate Streptococcus mutans exoenzyme GtfB binding to modulate cross-kingdom biofilm development in vivo. PLOS Pathog. 13, e1006407 (2017).
Pidwill, G. R., Rego, S., Jenkinson, H. F., Lamont, R. J. & Nobbs, A. H. Coassociation between group B streptococcus and Candida albicans promotes interactions with vaginal epithelium. Infect. Immun. 86, e00669–17 (2018).
Hoarau, G. et al. Bacteriome and mycobiome interactions underscore microbial dysbiosis in familial Crohn’s disease. MBio 7, e01250–16 (2016).
Trunk, K. et al. The type VI secretion system deploys antifungal effectors against microbial competitors. Nat. Microbiol. 3, 920–931 (2018).
Centeno, A., Davis, C. P., Cohen, M. S. & Warren, M. M. Modulation of Candida albicans attachment to human epithelial cells by bacteria and carbohydrates. Infect. Immun. 39, 1354–1360 (1983).
Levison, M. E. & Pitsakis, P. G. Susceptibility to experimental Candida albicans urinary tract infection in the rat. J. Infect. Dis. 155, 841–846 (1987).
Makrides, H. C. & MacFarlane, T. W. An investigation of the factors involved in increased adherence of C. albicans to epithelial cells mediated by E. coli. Microbios 38, 177–185 (1983).
Ponomarova, O. et al. Yeast creates a niche for symbiotic lactic acid bacteria through nitrogen overflow. Cell Syst. 5, 345–357 (2017).
Li, S. et al. The opportunistic human fungal pathogen Candida albicans promotes the growth and proliferation of commensal Escherichia coli through an iron-responsive pathway. Microbiol. Res. 207, 232–239 (2018).
Kong, E. F., Tsui, C., Kucharikova, S., Van Dijck, P. & Jabra-Rizk, M. A. Modulation of Staphylococcus aureus response to antimicrobials by the Candida albicans quorum sensing molecule farnesol. Antimicrob. Agents Chemother. 61, e01573–17 (2017).
Siavoshi, F. & Saniee, P. Vacuoles of Candida yeast as a specialized niche for Helicobacter pylori. World J. Gastroenterol. 20, 5263–5273 (2014).
van Leeuwen, P. T. et al. Interspecies Interactions between Clostridium difficile and Candida albicans. mSphere 1, e00187–16 (2016).
Lambooij, J. M., Hoogenkamp, M. A., Brandt, B. W., Janus, M. M. & Krom, B. P. Fungal mitochondrial oxygen consumption induces the growth of strict anaerobic bacteria. Fungal Genet. Biol. 109, 1–6 (2017).
Sovran, B. et al. Enterobacteriaceae are essential for the modulation of colitis severity by fungi. Microbiome 6, 152 (2018).
Peleg, A. Y., Hogan, D. A. & Mylonakis, E. Medically important bacterial-fungal interactions. Nat. Rev. Microbiol. 8, 340–349 (2010).
Seelig, M. S. Mechanisms by which antibiotics increase the incidence and severity of candidiasis and alter the immunological defenses. Bacteriol. Rev. 30, 442–459 (1966).
Samonis, G. et al. Prospective evaluation of effects of broad-spectrum antibiotics on gastrointestinal yeast colonization of humans. Antimicrob. Agents Chemother. 37, 51–53 (1993).
Garcia, C. et al. The human gut microbial metabolome modulates fungal growth via the TOR signaling pathway. mSphere 2, e00555–17 (2017).
Allonsius, C. N. et al. Interplay between Lactobacillus rhamnosus GG and Candida and the involvement of exopolysaccharides. Microb. Biotechnol. 10, 1753–1763 (2017).
Graham, C. E., Cruz, M. R., Garsin, D. A. & Lorenz, M. C. Enterococcus faecalis bacteriocin EntV inhibits hyphal morphogenesis, biofilm formation, and virulence of Candida albicans. Proc. Natl Acad. Sci. USA 114, 4507–4512 (2017).
Kim, Y. & Mylonakis, E. Killing of Candida albicans filaments by Salmonella enterica serovar Typhimurium is mediated by sopB effectors, parts of a type III secretion system. Eukaryot. Cell 10, 782–790 (2011).
Mayer, F. L. & Kronstad, J. W. Disarming fungal pathogens: Bacillus safensis inhibits virulence factor production and biofilm formation by Cryptococcus neoformans and Candida albicans. MBio 8, e01537–17 (2017).
Wagner, R. D. et al. Biotherapeutic effects of probiotic bacteria on candidiasis in immunodeficient mice. Infect. Immun. 65, 4165–4172 (1997).
Liang, W. et al. Lactic acid bacteria differentially regulate filamentation in two heritable cell types of the human fungal pathogen Candida albicans. Mol. Microbiol. 102, 506–519 (2016).
Cruz, M. R., Graham, C. E., Gagliano, B. C., Lorenz, M. C. & Garsin, D. A. Enterococcus faecalis inhibits hyphal morphogenesis and virulence of Candida albicans. Infect. Immun. 81, 189–200 (2013).
Cugini, C. et al. Farnesol, a common sesquiterpene, inhibits PQS production in Pseudomonas aeruginosa. Mol. Microbiol. 65, 896–906 (2007).
Leonhardt, I. et al. The fungal quorum-sensing molecule farnesol activates innate immune cells but suppresses cellular adaptive immunity. MBio 6, e00143 (2015).
Jabra-Rizk, M. A., Meiller, T. F., James, C. E. & Shirtliff, M. E. Effect of farnesol on Staphylococcus aureus biofilm formation and antimicrobial susceptibility. Antimicrob. Agents Chemother. 50, 1463–1469 (2006).
Jang, J. E. et al. The effect of rice with Aspergillus terreus on lipid metabolism in rats. Korean J. Food Sci. Technol. 47, 658–666 (2015).
Sokol, H. et al. Fungal microbiota dysbiosis in IBD. Gut 66, 1039–1048 (2017).
Everard, A., Matamoros, S., Geurts, L., Delzenne, N. M. & Cani, P. D. Saccharomyces boulardii administration changes gut microbiota and reduces hepatic steatosis, low-grade inflammation, and fat mass in obese and type 2 diabetic db/db mice. MBio 5, e01011–14 (2014).
Iliev, I. D. & Leonardi, I. Fungal dysbiosis: immunity and interactions at mucosal barriers. Nat. Rev. Immunol. 17, 635–646 (2017).
Netea, M. G., Joosten, L. A., van der Meer, J. W., Kullberg, B. J. & van de Veerdonk, F. L. Immune defence against Candida fungal infections. Nat. Rev. Immunol. 15, 630–642 (2015).
Underhill, D. M. & Iliev, I. D. The mycobiota: interactions between commensal fungi and the host immune system. Nat. Rev. Immunol. 14, 405–416 (2014).
Underhill, D. M. & Pearlman, E. Immune interactions with pathogenic and commensal fungi: a two-way street. Immunity 43, 845–858 (2015).
Wheeler, M. L. et al. Immunological consequences of intestinal fungal dysbiosis. Cell Host Microbe 19, 865–873 (2016).
Bacher, P. et al. Antigen-specific expansion of human regulatory T cells as a major tolerance mechanism against mucosal fungi. Mucosal Immunol. 7, 916–928 (2014).
Bedke, T. et al. Distinct and complementary roles for Aspergillus fumigatus-specific Tr1 and Foxp3 + regulatory T cells in humans and mice. Immunol. Cell Biol. 92, 659–670 (2014).
Bourgeois, C. & Kuchler, K. Fungal pathogens-a sweet and sour treat for toll-like receptors. Front. Cell. Infect. Microbiol. 2, 142 (2012).
Plato, A., Hardison, S. E. & Brown, G. D. Pattern recognition receptors in antifungal immunity. Semin. Immunopathol. 37, 97–106 (2015).
Eliaz, I. The role of galectin-3 as a marker of cancer and inflammation in a stage IV ovarian cancer patient with underlying pro-inflammatory comorbidities. Case Rep. Oncol. 6, 343–349 (2013).
Srivatsan, V., George, M. & Shanmugam, E. Utility of galectin-3 as a prognostic biomarker in heart failure: where do we stand? Eur. J. Prev. Cardiol. 22, 1096–1110 (2015).
Linden, J. R., Kunkel, D., Laforce-Nesbitt, S. S. & Bliss, J. M. The role of galectin-3 in phagocytosis of Candida albicans and Candida parapsilosis by human neutrophils. Cell. Microbiol. 15, 1127–1142 (2013).
Vautier, S., MacCallum, D. M. & Brown, G. D. C-Type lectin receptors and cytokines in fungal immunity. Cytokine 58, 89–99 (2012).
Li, S. S. et al. Identification of the fungal ligand triggering cytotoxic PRR-mediated NK cell killing of Cryptococcus and Candida. Nat. Commun. 9, 751 (2018).
Leonardi, I. et al. CX3CR1(+) mononuclear phagocytes control immunity to intestinal fungi. Science 359, 232–236 (2018).
Zhang, I., Pletcher, S. D., Goldberg, A. N., Barker, B. M. & Cope, E. K. Fungal microbiota in chronic airway inflammatory disease and emerging relationships with the host immune response. Front. Microbiol. 8, 2477 (2017).
Zhang, Z. et al. Peripheral lymphoid volume expansion and maintenance are controlled by gut microbiota via RALDH+dendritic cells. Immunity 44, 330–342 (2016).
Fujimura, K. E. et al. Neonatal gut microbiota associates with childhood multisensitized atopy and T cell differentiation. Nat. Med. 22, 1187–1191 (2016).
Fan, D. et al. Activation of HIF-1alpha and LL-37 by commensal bacteria inhibits Candida albicans colonization. Nat. Med. 21, 808–814 (2015).
Zelante, T. et al. Tryptophan catabolites from microbiota engage aryl hydrocarbon receptor and balance mucosal reactivity via interleukin-22. Immunity 39, 372–385 (2013).
Quinton, J. F. et al. Anti-Saccharomyces cerevisiae mannan antibodies combined with antineutrophil cytoplasmic autoantibodies in inflammatory bowel disease: prevalence and diagnostic role. Gut 42, 788–791 (1998).
Muller, S. et al. Mannan-binding lectin deficiency results in unusual antibody production and excessive experimental colitis in response to mannose-expressing mild gut pathogens. Gut 59, 1493–1500 (2010).
Standaert-Vitse, A. et al. Candida albicans is an immunogen for anti-Saccharomyces cerevisiae antibody markers of Crohn’s disease. Gastroenterology 130, 1764–1775 (2006).
McFarland, L. V. Systematic review and meta-analysis of Saccharomyces boulardii in adult patients. World J. Gastroenterol. 16, 2202–2222 (2010).
Jawhara, S. et al. Colonization of mice by Candida albicans is promoted by chemically induced colitis and augments inflammatory responses through galectin-3. J. Infect. Dis. 197, 972–980 (2008).
Standaert-Vitse, A. et al. Candida albicans colonization and ASCA in familial Crohn’s disease. Am. J. Gastroenterol. 104, 1745–1753 (2009).
Jostins, L. et al. Host-microbe interactions have shaped the genetic architecture of inflammatory bowel disease. Nature 491, 119–124 (2012).
Mathew, C. G. New links to the pathogenesis of Crohn disease provided by genome-wide association scans. Nat. Rev. Genet. 9, 9–14 (2008).
Iliev, I. D. et al. Interactions between commensal fungi and the C-type lectin receptor Dectin-1 influence colitis. Science 336, 1314–1317 (2012).
Sokol, H. et al. Card9 mediates intestinal epithelial cell restitution, T-helper 17 responses, and control of bacterial infection in mice. Gastroenterology 145, 591–601 (2013).
El Mouzan, M. et al. Fungal microbiota profile in newly diagnosed treatment-naive children with Crohn’s disease. J. Crohns Colitis 11, 586–592 (2017).
Liguori, G. et al. Fungal dysbiosis in mucosa-associated microbiota of Crohn’s disease patients. J. Crohns Colitis 10, 296–305 (2016).
Mukhopadhya, I. et al. The fungal microbiota of de-novo paediatric inflammatory bowel disease. Microbes Infect. 17, 304–310 (2015).
Chehoud, C. et al. Fungal signature in the gut microbiota of pediatric patients with inflammatory bowel disease. Inflamm. Bowel Dis. 21, 1948–1956 (2015).
Qiu, X. et al. Changes in the composition of intestinal fungi and their role in mice with dextran sulfate sodium-induced colitis. Sci. Rep. 5, 10416 (2015).
Tang, C. et al. Inhibition of dectin-1 signaling ameliorates colitis by inducing Lactobacillus-mediated regulatory T cell expansion in the intestine. Cell Host Microbe 18, 183–197 (2015).
Iliev, I. D. Dectin-1 exerts dual control in the gut. Cell Host Microbe 18, 139–141 (2015).
Botschuijver, S. et al. Intestinal fungal dysbiosis is associated with visceral hypersensitivity in patients with irritable bowel syndrome and rats. Gastroenterology 153, 1026–1039 (2017).
Brennan, C. A. & Garrett, W. S. Gut microbiota, inflammation, and colorectal cancer. Annu. Rev. Microbiol. 70, 395–411 (2016).
Luan, C. et al. Dysbiosis of fungal microbiota in the intestinal mucosa of patients with colorectal adenomas. Sci. Rep. 5, 7980 (2015).
Richard, M. L. et al. Mucosa-associated microbiota dysbiosis in colitis associated cancer. Gut Microbes 9, 131–142 (2018).
Gao, R. et al. Dysbiosis signature of mycobiota in colon polyp and colorectal cancer. Eur. J. Clin. Microbiol. Infect. Dis. 36, 2457–2468 (2017).
Yang, A. M. et al. Intestinal fungi contribute to development of alcoholic liver disease. J. Clin. Invest. 127, 2829–2841 (2017).
Marchesi, J. R. & Ravel, J. The vocabulary of microbiome research: a proposal. Microbiome 3, 31 (2015).
Lachance, M. A., Gilbert, D. G. & Starmer, W. T. Yeast communities associated with Drosophila species and related flies in an eastern oak-pine forest: a comparison with western communities. J. Ind. Microbiol. 14, 484–494 (1995).
Author information
Authors and Affiliations
Contributions
Both authors researched data for the article, made substantial contributions to discussion of the article content and wrote and reviewed and/or edited the manuscript before submission.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Related links
European MetaHit project: http://www.metahit.eu/
International Nucleotide Sequence Database: http://www.insdc.org
Rights and permissions
About this article
Cite this article
Richard, M.L., Sokol, H. The gut mycobiota: insights into analysis, environmental interactions and role in gastrointestinal diseases. Nat Rev Gastroenterol Hepatol 16, 331–345 (2019). https://doi.org/10.1038/s41575-019-0121-2
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41575-019-0121-2
This article is cited by
-
Insights into gut microbiomes in stem cell transplantation by comprehensive shotgun long-read sequencing
Scientific Reports (2024)
-
Resveratrol alleviates DSS-induced IBD in mice by regulating the intestinal microbiota-macrophage-arginine metabolism axis
European Journal of Medical Research (2023)
-
Biogeography, succession, and origin of the chicken intestinal mycobiome
Microbiome (2022)
-
The gut metagenomics and metabolomics signature in patients with inflammatory bowel disease
Gut Pathogens (2022)
-
Differences in the Intestinal Flora of Patients with Inflammatory Bowel Disease in Southwest China
Indian Journal of Microbiology (2022)