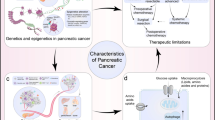
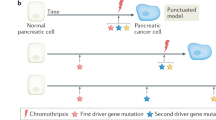
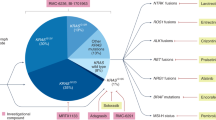
Abstract
Cancers that appear morphologically similar often have dramatically different clinical features, respond variably to therapy and have a range of outcomes. Compelling evidence now demonstrates that differences in the molecular pathology of otherwise indistinguishable cancers substantially impact the clinical characteristics of the disease. Molecular subtypes now guide preclinical and clinical therapeutic development and treatment in many cancer types. The ability to predict optimal therapeutic strategies ahead of treatment improves overall patient outcomes, minimizing treatment-related morbidity and cost. Although clinical decision making based on histopathological criteria underpinned by robust data is well established in many cancer types, subtypes of pancreatic cancer do not currently inform treatment decisions. However, accumulating molecular data are defining subgroups in pancreatic cancer with distinct biology and potential subtype-specific therapeutic vulnerabilities, providing the opportunity to define a de novo clinically applicable molecular taxonomy. This Review summarizes current knowledge concerning the molecular subtyping of pancreatic cancer and explores future strategies for using a molecular taxonomy to guide therapeutic development and ultimately routine therapy with the overall goal of improving outcomes for this disease.
Key points
-
Pancreatic cancer is soon to become the second leading cause of cancer-related death.
-
Histopathological criteria do not adequately inform treatment decisions for pancreatic cancer.
-
A molecular taxonomy could improve outcomes with current treatments and accelerate therapeutic development through better patient selection.
-
Emerging molecular taxonomies define biological differences between subtypes that are associated with prognosis.
-
Genomic and transcriptomic subtypes potentially enrich for therapeutic vulnerabilities and require preclinical and clinical assessment.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Hudson, T. J. et al. International network of cancer genome projects. Nature 464, 993–998 (2010).
The Cancer Genome Atlas Research Network. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature 455, 1061–1068 (2008).
Biankin, A. V. & Hudson, T. J. Somatic variation and cancer: therapies lost in the mix. Hum. Genet. 130, 79–91 (2011).
Biankin, A. V., Piantadosi, S. & Hollingsworth, S. J. Patient-centric trials for therapeutic development in precision oncology. Nature 526, 361–370 (2015).
Hollingsworth, S. J. & Biankin, A. V. The challenges of precision oncology drug development and implementation. Public Health Genomics 18, 338–348 (2015).
Swanton, C. et al. Consensus on precision medicine for metastatic cancers: a report from the MAP conference. Ann. Oncol. 27, 1443–1448 (2016).
American Cancer Society. Key statistics for pancreatic cancer. Cancer.org https://www.cancer.org/cancer/pancreatic-cancer/about/key-statistics.html (2018).
Siegel, R. L., Miller, K. D. & Jemal, A. Cancer statistics, 2017. CA Cancer J. Clin. 67, 7–30 (2017).
Rahib, L. et al. Projecting cancer incidence and deaths to 2030: the unexpected burden of thyroid, liver, and pancreas cancers in the United States. Cancer Res. 74, 2913–2921 (2014).
National Cancer Institute SEER. Cancer stat facts: common cancer sites. SEER https://seer.cancer.gov/statfacts/html/common.html (2018).
Ryan, D. P., Hong, T. S. & Bardeesy, N. Pancreatic adenocarcinoma. N. Engl. J. Med. 371, 2140–2141 (2014).
The Cancer Genome Atlas Research Network. Comprehensive molecular profiling of lung adenocarcinoma. Nature 511, 543–550 (2014).
Alexandrov, L. B. et al. Signatures of mutational processes in human cancer. Nature 500, 415–421 (2013).
Petersen, G. M. Familial pancreatic cancer. Semin. Oncol. 43, 548–553 (2016).
Klein, A. P. et al. Prospective risk of pancreatic cancer in familial pancreatic cancer kindreds. Cancer Res. 64, 2634–2638 (2004).
Brune, K. A. et al. Importance of age of onset in pancreatic cancer kindreds. J. Natl Cancer Inst. 102, 119–126 (2010).
Jones, S. et al. Exomic sequencing identifies PALB2 as a pancreatic cancer susceptibility gene. Science 324, 217 (2009).
Humphris, J. L. et al. Clinical and pathologic features of familial pancreatic cancer. Cancer 120, 3669–3675 (2014).
Wolpin, B. M. et al. ABO blood group and the risk of pancreatic cancer. J. Natl Cancer Inst. 101, 424–431 (2009).
Hu, Z. I. et al. Evaluating mismatch repair deficiency in pancreatic adenocarcinoma: challenges and recommendations. Clin. Cancer Res. 24, 1326–1336 (2018).
Childs, E. J. et al. Association of common susceptibility variants of pancreatic cancer in higher-risk patients: a PACGENE study. Cancer Epidemiol. Biomarkers Prev. 25, 1185–1191 (2016).
Grant, R. C. et al. Prevalence of germline mutations in cancer predisposition genes in patients with pancreatic cancer. Gastroenterology 148, 556–564 (2014).
Shindo, K. et al. Deleterious germline mutations in patients with apparently sporadic pancreatic adenocarcinoma. J. Clin. Oncol. 35, 3382–3390 (2017).
Humphris, J., Chang, D. K. & Biankin, A. V. Inherited susceptibility to pancreatic cancer in the era of next-generation sequencing. Gastroenterology 148, 496–498 (2015).
Yurgelun, M. B. et al. Germline cancer susceptibility gene variants, somatic second hits, and survival outcomes in patients with resected pancreatic cancer. Genet. Med. 21, 213–223 (2019).
Canto, M. I. et al. International Cancer of the Pancreas Screening (CAPS) Consortium summit on the management of patients with increased risk for familial pancreatic cancer. Gut 62, 339–347 (2013).
Langer, P. et al. Five years of prospective screening of high-risk individuals from families with familial pancreatic cancer. Gut 58, 1410–1418 (2009).
McWilliams, R. R. et al. Risk factors for early-onset and very-early-onset pancreatic adenocarcinoma: a pancreatic cancer case-control consortium (PanC4) analysis. Pancreas 45, 311–316 (2016).
Kloppel, G. & Luttges, J. WHO-classification 2000: exocrine pancreatic tumors. Verh. Dtsch. Ges. Pathol. 85, 219–228 (2001).
Kardon, D. E., Thompson, L. D., Przygodzki, R. M. & Heffess, C. S. Adenosquamous carcinoma of the pancreas: a clinicopathologic series of 25 cases. Mod. Pathol. 14, 443–451 (2001).
Basturk, O. et al. Poorly differentiated neuroendocrine carcinomas of the pancreas: a clinicopathologic analysis of 44 cases. Am. J. Surg. Pathol. 38, 437–447 (2014).
Basturk, O. et al. A revised classification system and recommendations from the Baltimore Consensus Meeting for neoplastic precursor lesions in the pancreas. Am. J. Surg. Pathol. 39, 1730–1741 (2015).
Hruban, R. H. et al. An illustrated consensus on the classification of pancreatic intraepithelial neoplasia and intraductal papillary mucinous neoplasms. Am. J. Surg. Pathol. 28, 977–987 (2004).
Brugge, W. R., Lauwers, G. Y., Sahani, D., Fernandez-del Castillo, C. & Warshaw, A. L. Cystic neoplasms of the pancreas. N. Engl. J. Med. 351, 1218–1226 (2004).
Balachandran, V. P. et al. Identification of unique neoantigen qualities in long-term survivors of pancreatic cancer. Nature 551, 512–516 (2017).
Scarpa, A., Real, F. X. & Luchini, C. Genetic unrelatedness of co-occurring pancreatic adenocarcinomas and IPMNs challenges current views of clinical management. Gut 67, 1561–1563 (2018).
Springer, S. et al. A combination of molecular markers and clinical features improve the classification of pancreatic cysts. Gastroenterology 149, 1501–1510 (2015).
Reid, M. D., Bagci, P. & Adsay, N. V. Histopathologic assessment of pancreatic cancer: does one size fit all? J. Surg. Oncol. 107, 67–77 (2013).
Pishvaian, M. J. & Brody, J. R. Therapeutic implications of molecular subtyping for pancreatic cancer. Oncology 31, 159–166 (2017).
Biankin, A. V. et al. Expression of S100A2 calcium-binding protein predicts response to pancreatectomy for pancreatic cancer. Gastroenterology 137, 558–568 (2009).
FDA News Release. FDA approves first cancer treatment for any solid tumor with a specific genetic feature. FDA.gov https://www.fda.gov/newsevents/newsroom/pressannouncements/ucm560167.htm (2017).
Humphris, J. L. et al. Hypermutation in pancreatic cancer. Gastroenterology 152, 68–74 (2017).
Niu, B. et al. MSIsensor: microsatellite instability detection using paired tumor-normal sequence data. Bioinformatics 30, 1015–1016 (2014).
Garcea, G., Neal, C. P., Pattenden, C. J., Steward, W. P. & Berry, D. P. Molecular prognostic markers in pancreatic cancer: a systematic review. Eur. J. Cancer 41, 2213–2236 (2005).
Iacobuzio-Donahue, C. A. et al. DPC4 gene status of the primary carcinoma correlates with patterns of failure in patients with pancreatic cancer. J. Clin. Oncol. 27, 1806–1813 (2009).
Dreyer, S. B. et al. Precision oncology in surgery: patient selection biomarkers for operable pancreatic cancer [abstract 10]. Eur. J. Surg. Oncol. 44, 1838 (2017).
Humphris, J. L. et al. The prognostic and predictive value of serum CA19.9 in pancreatic cancer. Ann. Oncol. 23, 1713–1722 (2012).
Aguirre, A. J. et al. Real-time genomic characterization of advanced pancreatic cancer to enable precision medicine. Cancer Discov. 8, 1096–1111 (2018).
Qian, Z. R. et al. Association of alterations in main driver genes with outcomes of patients with resected pancreatic ductal adenocarcinoma. JAMA Oncol. 4, e173420 (2018).
Smit, V. T. et al. KRAS codon 12 mutations occur very frequently in pancreatic adenocarcinomas. Nucleic Acids Res. 16, 7773–7782 (1988).
Jones, S. et al. Somatic mutations in the chromatin remodeling gene ARID1A occur in several tumor types. Hum. Mut. 33, 100–103 (2012).
Jones, S. et al. Core signaling pathways in human pancreatic cancers revealed by global genomic analyses. Science 321, 1801–1806 (2008).
Biankin, A. V. et al. Pancreatic cancer genomes reveal aberrations in axon guidance pathway genes. Nature 491, 399–405 (2012).
Chang, D. K., Grimmond, S. M. & Biankin, A. V. Pancreatic cancer genomics. Curr. Opin. Genet. Dev. 24, 74–81 (2014).
Waddell, N. et al. Whole genomes redefine the mutational landscape of pancreatic cancer. Nature 518, 495–501 (2015).
Bailey, P. et al. Genomic analyses identify molecular subtypes of pancreatic cancer. Nature 531, 47–52 (2016).
Dreyer, S. B., Chang, D. K., Bailey, P. & Biankin, A. V. Pancreatic cancer genomes: implications for clinical management and therapeutic development. Clin. Cancer Res. 23, 1638–1646 (2017).
Moffitt, R. A. et al. Virtual microdissection identifies distinct tumor- and stroma-specific subtypes of pancreatic ductal adenocarcinoma. Nat. Genet. 47, 1168–1178 (2015).
Witkiewicz, A. K. et al. Whole-exome sequencing of pancreatic cancer defines genetic diversity and therapeutic targets. Nat. Commun. 6, 6744 (2015).
The Cancer Genome Atlas Research Network. Integrated genomic characterization of pancreatic ductal adenocarcinoma. Cancer Cell 32, 185–203 (2017).
Cowley, M. J. et al. Understanding pancreatic cancer genomes. J. Hepatobiliary Pancreat. Sci. 20, 549–556 (2013).
Kleeff, J. et al. Pancreatic cancer. Nat. Rev. Dis. Primers 2, 16022 (2016).
Slamon, D. J. et al. Use of chemotherapy plus a monoclonal antibody against HER2 for metastatic breast cancer that overexpresses HER2. N. Engl. J. Med. 344, 783–792 (2001).
Bollag, G. et al. Clinical efficacy of a RAF inhibitor needs broad target blockade in BRAF-mutant melanoma. Nature 467, 596–599 (2010).
Planchard, D. et al. Dabrafenib plus trametinib in patients with previously untreated BRAF(V600E)-mutant metastatic non-small-cell lung cancer: an open-label, phase 2 trial. Lancet Oncol. 18, 1307–1316 (2017).
Ledermann, J. et al. Olaparib maintenance therapy in platinum-sensitive relapsed ovarian cancer. N. Engl. J. Med. 366, 1382–1392 (2012).
Morran, D. C. et al. Targeting mTOR dependency in pancreatic cancer. Gut 63, 1481–1489 (2014).
Weissmueller, S. et al. Mutant p53 drives pancreatic cancer metastasis through cell-autonomous PDGF receptor beta signaling. Cell 157, 382–394 (2014).
Miller, B. W. et al. Targeting the LOX/hypoxia axis reverses many of the features that make pancreatic cancer deadly: inhibition of LOX abrogates metastasis and enhances drug efficacy. EMBO Mol. Med. 7, 1063–1076 (2015).
Chou, A. et al. Clinical and molecular characterization of HER2 amplified-pancreatic cancer. Genome Med. 5, 78 (2013).
Chmielecki, J. et al. Comprehensive genomic profiling of pancreatic acinar cell carcinomas identifies recurrent RAF fusions and frequent inactivation of DNA repair genes. Cancer Discov. 4, 1398–1405 (2014).
Lowery, M. A. et al. Real-time genomic profiling of pancreatic ductal adenocarcinoma: potential actionability and correlation with clinical phenotype. Clin. Cancer Res. 23, 6094–6100 (2017).
Foster, S. A. et al. Activation mechanism of oncogenic deletion mutations in BRAF, EGFR, and HER2. Cancer Cell 29, 477–493 (2016).
Chang, D. K., Grimmond, S. M., Evans, T. R. J. & Biankin, A. V. Mining the genomes of exceptional responders. Nat. Rev. Cancer 14, 291–292 (2014).
Lord, C. J. & Ashworth, A. The DNA damage response and cancer therapy. Nature 481, 287–294 (2012).
McBride, D. J. et al. Tandem duplication of chromosomal segments is common in ovarian and breast cancer genomes. J. Pathol. 227, 446–455 (2012).
Polak, P. et al. A mutational signature reveals alterations underlying deficient homologous recombination repair in breast cancer. Nat. Genet. 49, 1476–1486 (2017).
Davies, H. et al. HRDetect is a predictor of BRCA1 and BRCA2 deficiency based on mutational signatures. Nat. Med. 23, 517–525 (2017).
Bamford, S. et al. The COSMIC (Catalogue of Somatic Mutations in Cancer) database and website. Br. J. Cancer 91, 355–358 (2004).
Forbes, S. A. et al. COSMIC: mining complete cancer genomes in the Catalogue of Somatic Mutations in Cancer. Nucleic Acids Res. 39, D945–D950 (2011).
Catalogue of Somatic Mutations in Cancer. Signatures of mutational processes in human cancer. COSMIC https://cancer.sanger.ac.uk/cosmic/signatures (2017).
Swisher, E. M. et al. Rucaparib in relapsed, platinum-sensitive high-grade ovarian carcinoma (ARIEL2 Part 1): an international, multicentre, open-label, phase 2 trial. Lancet Oncol. 18, 75–87 (2017).
Alizadeh, A. A. et al. Distinct types of diffuse large B cell lymphoma identified by gene expression profiling. Nature 403, 503–511 (2000).
Perou, C. M. et al. Molecular portraits of human breast tumours. Nature 406, 747–752 (2000).
Sadanandam, A. et al. A colorectal cancer classification system that associates cellular phenotype and responses to therapy. Nat. Med. 19, 619–625 (2013).
De Sousa, E. M. F. et al. Poor-prognosis colon cancer is defined by a molecularly distinct subtype and develops from serrated precursor lesions. Nat. Med. 19, 614–618 (2013).
Marisa, L. et al. Gene expression classification of colon cancer into molecular subtypes: characterization, validation, and prognostic value. PLOS Med. 10, e1001453 (2013).
Guinney, J. et al. The consensus molecular subtypes of colorectal cancer. Nat. Med. 21, 1350–1356 (2015).
Collisson, E. A. et al. Subtypes of pancreatic ductal adenocarcinoma and their differing responses to therapy. Nat. Med. 17, 500–503 (2011).
Decker, K., Goldman, D. C., Grasch, C. L. & Sussel, L. Gata6 is an important regulator of mouse pancreas development. Dev. Biol. 298, 415–429 (2006).
Jamieson, N. B., Chang, D. K. & Biankin, A. V. Cancer genetics and implications for clinical management. Surg. Clin. North Am. 95, 919–934 (2015).
Vogelstein, B. et al. Cancer genome landscapes. Science 339, 1546–1558 (2013).
Kassahn, K. S. et al. Somatic point mutation calling in low cellularity tumors. PLOS ONE 8, e74380 (2013).
Song, S. et al. qpure: a tool to estimate tumor cellularity from genome-wide single-nucleotide polymorphism profiles. PLOS ONE 7, e45835 (2012).
Nones, K. et al. Genome-wide DNA methylation patterns in pancreatic ductal adenocarcinoma reveal epigenetic deregulation of SLIT-ROBO, ITGA2 and MET signaling. Int. J. Cancer 135, 1110–1118 (2014).
Hoadley, K. A. et al. Multiplatform analysis of 12 cancer types reveals molecular classification within and across tissues of origin. Cell 158, 929–944 (2014).
Biankin, A. V. & Maitra, A. Subtyping pancreatic cancer. Cancer Cell 28, 411–413 (2015).
Puleo, F. et al. Stratification of pancreatic ductal adenocarcinomas based on tumor and microenvironment features. Gastroenterology 155, 1999–2013 (2018).
Noll, E. M. et al. CYP3A5 mediates basal and acquired therapy resistance in different subtypes of pancreatic ductal adenocarcinoma. Nat. Med. 22, 278–287 (2016).
Knudsen, E. S. et al. Pancreatic cancer cell lines as patient-derived avatars: genetic characterisation and functional utility. Gut 67, 508–520 (2017).
Kopp, J. L. et al. Identification of Sox9-dependent acinar-to-ductal reprogramming as the principal mechanism for initiation of pancreatic ductal adenocarcinoma. Cancer Cell 22, 737–750 (2012).
Morris, J. P. t., Cano, D. A., Sekine, S., Wang, S. C. & Hebrok, M. Beta-catenin blocks Kras-dependent reprogramming of acini into pancreatic cancer precursor lesions in mice. J. Clin. Invest. 120, 508–520 (2010).
Steele, C. W. et al. CXCR2 inhibition profoundly suppresses metastases and augments immunotherapy in pancreatic ductal adenocarcinoma. Cancer Cell 29, 832–845 (2016).
Candido, J. B. et al. CSF1R(+) macrophages sustain pancreatic tumor growth through T cell suppression and maintenance of key gene programs that define the squamous subtype. Cell Rep. 23, 1448–1460 (2018).
Xu, Z. et al. Role of pancreatic stellate cells in pancreatic cancer metastasis. Am. J. Pathol. 177, 2585–2596 (2010).
Zhao, X. et al. Single-cell RNA-seq reveals a distinct transcriptome signature of aneuploid hematopoietic cells. Blood 130, 2762–2773 (2017).
Habib, N. et al. Massively parallel single-nucleus RNA-seq with DroNc-seq. Nat. Methods 14, 955–958 (2017).
Sinha, S. et al. PanIN neuroendocrine cells promote tumorigenesis via neuronal cross-talk. Cancer Res. 77, 1868–1879 (2017).
Delgiorno, K. E. et al. Identification and manipulation of biliary metaplasia in pancreatic tumors. Gastroenterology 146, 233–244 (2014).
Andricovich, J. et al. Loss of KDM6A activates super-enhancers to induce gender-specific squamous-like pancreatic cancer and confers sensitivity to BET inhibitors. Cancer Cell 33, 512–526 (2018).
Sivakumar, S., de Santiago, I., Chlon, L. & Markowetz, F. Master regulators of oncogenic KRAS response in pancreatic cancer: an integrative network biology analysis. PLOS Med. 14, e1002223 (2017).
Kim, S. T. et al. Correlating programmed death ligand 1 (PD-L1) expression, mismatch repair deficiency, and outcomes across tumor types: implications for immunotherapy. Oncotarget 8, 77415–77423 (2017).
Iacobuzio-Donahue, C. A. Genetic evolution of pancreatic cancer: lessons learnt from the pancreatic cancer genome sequencing project. Gut 61, 1085–1094 (2011).
Sequist, L. V. et al. Genotypic and histological evolution of lung cancers acquiring resistance to EGFR inhibitors. Sci. Transl Med. 3, 75ra26 (2011).
Furukawa, T. et al. Whole exome sequencing reveals recurrent mutations in BRCA2 and FAT genes in acinar cell carcinomas of the pancreas. Sci. Rep. 5, 8829 (2015).
Jiao, Y. et al. Whole-exome sequencing of pancreatic neoplasms with acinar differentiation. J. Pathol. 232, 428–435 (2014).
Hall, J. C. et al. Novel patient-derived xenograft mouse model for pancreatic acinar cell carcinoma demonstrates single agent activity of oxaliplatin. J. Transl Med. 14, 129 (2016).
Botton, T. et al. Recurrent BRAF kinase fusions in melanocytic tumors offer an opportunity for targeted therapy. Pigment Cell Melanoma Res. 26, 845–851 (2013).
Menzies, A. M. et al. Clinical activity of the MEK inhibitor trametinib in metastatic melanoma containing BRAF kinase fusion. Pigment Cell Melanoma Res. 28, 607–610 (2015).
Agaimy, A. et al. Pancreatic undifferentiated rhabdoid carcinoma: KRAS alterations and SMARCB1 expression status define two subtypes. Mod. Pathol. 28, 248–260 (2015).
Connor, A. A. et al. Association of distinct mutational signatures with correlates of increased immune activity in pancreatic ductal adenocarcinoma. JAMA Oncol. 3, 774–783 (2017).
Aung, K. L. et al. Genomics-driven precision medicine for advanced pancreatic cancer — early results from the COMPASS trial. Clin. Cancer Res. 24, 1344–1354 (2017).
de Santiago, I. et al. Immuno-phenotypes of pancreatic ductal adenocarcinoma: metaanalysis of transcriptional subtypes. Preprint at https://www.biorxiv.org/content/early/2017/10/05/198903 (2017).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Related links
Accelerating Research in Genomic Oncology: http://www.icgcargo.org/
Enhanced Pancreatic Cancer Profiling for Individualized Care (EPPIC): https://www.tfri.ca/en/NewsEvents/news/news-releases-detail/2018/03/06/canadian-pancreatic-cancer-research-team-provides-personalized-medicine-new-hope-to-patients
International Cancer Genome Consortium: http://www.icgc.org/
Precision-Panc: http://www.precisionpanc.org/
Precision Promise: http://www.pancan.org/research/precision-promise/
Rights and permissions
About this article
Cite this article
Collisson, E.A., Bailey, P., Chang, D.K. et al. Molecular subtypes of pancreatic cancer. Nat Rev Gastroenterol Hepatol 16, 207–220 (2019). https://doi.org/10.1038/s41575-019-0109-y
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41575-019-0109-y
This article is cited by
-
PancrESS – a meta-analysis resource for understanding cell-type specific expression in the human pancreas
BMC Genomics (2024)
-
Prolonged HPA axis dysregulation in postpartum depression associated with adverse early life experiences: a cross-species translational study
Nature Mental Health (2024)
-
Decoding the basis of histological variation in human cancer
Nature Reviews Cancer (2024)
-
Studying the impact of marital status on diagnosis and survival prediction in pancreatic ductal carcinoma using machine learning methods
Scientific Reports (2024)
-
Machine learning identifies SLC6A14 as a novel biomarker promoting the proliferation and metastasis of pancreatic cancer via Wnt/β-catenin signaling
Scientific Reports (2024)