Abstract
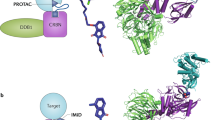
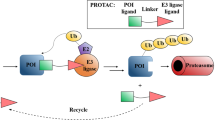
Proteolysis-targeting chimeras (PROTACs) are an emerging drug modality that may offer new opportunities to circumvent some of the limitations associated with traditional small-molecule therapeutics. By analogy with the concept of the ‘druggable genome’, the question arises as to which potential drug targets might PROTAC-mediated protein degradation be most applicable. Here, we present a systematic approach to the assessment of the PROTAC tractability (PROTACtability) of protein targets using a series of criteria based on data and information from a diverse range of relevant publicly available resources. Our approach could support decision-making on whether or not a particular target may be amenable to modulation using a PROTAC. Using our approach, we identified 1,067 proteins of the human proteome that have not yet been described in the literature as PROTAC targets that offer potential opportunities for future PROTAC-based efforts.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
The full output of the PROTACtability assessment pipeline is provided as an Excel spreadsheet (Supplementary Table 1); each row corresponds to a protein-coding gene and the columns represent the various bucket assessment results or data types. A more detailed description of the column headings can be found in Supplementary Box 1 and in the main text.
References
Chamberlain, P. P. & Hamann, L. G. Development of targeted protein degradation therapeutics. Nat. Chem. Biol. 15, 937–944 (2019).
Sun, X. et al. PROTACs: great opportunities for academia and industry. Signal Transduct. Target. Ther. 4, 64 (2019).
Ding, Y., Fei, Y. & Lu, B. Emerging new concepts of degrader technologies. Trends Pharmacol. Sci. 41, 464–474 (2020).
Pettersson, M. & Crews, C. M. PROteolysis TArgeting chimeras (PROTACs) — past, present and future. Drug Discov. Today Technol. 31, 15–27 (2019).
An, S. & Fu, L. Small-molecule PROTACs: An emerging and promising approach for the development of targeted therapy drugs. EBioMedicine 36, 553–562 (2018).
Nalawansha, D. A. & Crews, C. M. PROTACs: an emerging therapeutic modality in precision medicine. Cell Chem. Biol. 27, 998–1014 (2020).
Toure, M. & Crews, C. M. Small-molecule PROTACS: new approaches to protein degradation. Angew. Chem. Int. Ed. 55, 1966–1973 (2016).
Sakamoto, K. M. et al. Protacs: chimeric molecules that target proteins to the Skp1-Cullin-F box complex for ubiquitination and degradation. Proc. Natl Acad. Sci. USA 98, 8554–8559 (2001).
Nakayama, K. I. & Nakayama, K. Ubiquitin ligases: cell-cycle control and cancer. Nat. Rev. Cancer 6, 369–381 (2006).
Behrends, C. & Harper, J. W. Constructing and decoding unconventional ubiquitin chains. Nat. Struct. Mol. Biol. 18, 520–528 (2011).
Zheng, N. & Shabek, N. Ubiquitin ligases: structure, function, and regulation. Annu. Rev. Biochem. 86, 129–157 (2017).
Cromm, P. M., Samarasinghe, K. T. G., Hines, J. & Crews, C. M. Addressing kinase-independent functions of Fak via PROTAC-mediated degradation. J. Am. Chem. Soc. 140, 17019–17026 (2018).
Popow, J. et al. Highly selective PTK2 proteolysis targeting chimeras to probe focal adhesion kinase scaffolding functions. J. Med. Chem. 62, 2508–2520 (2019).
Song, Y. et al. Development and preclinical validation of a novel covalent ubiquitin receptor Rpn13 degrader in multiple myeloma. Leukemia 33, 2685–2694 (2019).
Bai, L. et al. A potent and selective small-molecule degrader of STAT3 achieves complete tumor regression in vivo. Cancer Cell 36, 498–511.e17 (2019).
Kaur, T., Menon, A. & Garner, A. L. Synthesis of 7-benzylguanosine cap-analogue conjugates for eIF4E targeted degradation. Eur. J. Med. Chem. 166, 339–350 (2019).
Bondeson, D. P. et al. Catalytic in vivo protein knockdown by small-molecule PROTACs. Nat. Chem. Biol. 11, 611–617 (2015).
Winter, G. E. et al. Phthalimide conjugation as a strategy for in vivo target protein degradation. Science 348, 1376–1381 (2015).
Zengerle, M., Chan, K.-H. & Ciulli, A. Selective small molecule induced degradation of the BET bromodomain protein BRD4. ACS Chem. Biol. 10, 1770–1777 (2015).
Konstantinidou, M. et al. PROTACs – a game-changing technology. Expert Opin. Drug Discov. 14, 1255–1268 (2019).
Wang, Y., Jiang, X., Feng, F., Liu, W. & Sun, H. Degradation of proteins by PROTACs and other strategies. Acta Pharm. Sin. B 10, 207–238 (2020).
Burslem, G. M. et al. The advantages of targeted protein degradation over inhibition: an RTK case study. Cell Chem. Biol. 25, 67–77.e3 (2018).
Mares, A. et al. Extended pharmacodynamic responses observed upon PROTAC-mediated degradation of RIPK2. Commun. Biol. 3, 140 (2020).
De Vita, E., Maneiro, M. & Tate, E. W. The missing link between (un)druggable and degradable KRAS. ACS Cent. Sci. 6, 1281–1284 (2020).
Snyder, L. B. et al. In Proc. 112th Annual Meeting Am. Assoc. Cancer Res. 10–15 Abstr. 43 (AACR, 2021).
Snyder, L. B. et al. In Proc. 112th Annual Meeting Am. Assoc. Cancer Res. 10–15 Abstr. 44 (AACR, 2021).
Gleeson, M. P., Hersey, A., Montanari, D. & Overington, J. Probing the links between in vitro potency, ADMET and physicochemical parameters. Nat. Rev. Drug Discov. 10, 197–208 (2011).
Keserü, G. M. & Makara, G. M. The influence of lead discovery strategies on the properties of drug candidates. Nat. Rev. Drug Discov. 8, 203–212 (2009).
Meanwell, N. A. Improving drug candidates by design: a focus on physicochemical properties as a means of improving compound disposition and safety. Chem. Res. Toxicol. 24, 1420–1456 (2011).
Leeson, P. D. & Springthorpe, B. The influence of drug-like concepts on decision-making in medicinal chemistry. Nat. Rev. Drug Discov. 6, 881–890 (2007).
Manallack, D. T., Prankerd, R. J., Yuriev, E., Oprea, T. I. & Chalmers, D. K. The significance of acid/base properties in drug discovery. Chem. Soc. Rev. 42, 485–496 (2013).
Pike, A., Williamson, B., Harlfinger, S., Martin, S. & McGinnity, D. F. Optimising proteolysis-targeting chimeras (PROTACs) for oral drug delivery: a drug metabolism and pharmacokinetics perspective. Drug Discov. Today 25, 1793–1800 (2020).
Hopkins, A. L. & Groom, C. R. The druggable genome. Nat. Rev. Drug Discov. 1, 727–730 (2002).
Gerry, C. J. & Schreiber, S. L. Unifying principles of bifunctional, proximity-inducing small molecules. Nat. Chem. Biol. 16, 369–378 (2020).
El-Ahmad, Y. et al. Discovery of 6-(2,4-dichlorophenyl)-5-[4-[(3S)-1-(3-fluoropropyl)pyrrolidin-3-yl]oxyphenyl]-8,9-dihydro-7H-benzo[7]annulene-2-carboxylic acid (SAR439859), a potent and selective estrogen receptor degrader (SERD) for the treatment of estrogen-receptor-positive breast cancer.J. Med. Chem. 63, 512–518 (2020).
Dauvois, S., Danielian, P. S., White, R. & Parker, M. G. Antiestrogen ICI 164,384 reduces cellular estrogen receptor content by increasing its turnover. Proc. Natl Acad. Sci. Usa. 89, 4037–4041 (1992).
Wu, Y.-L. et al. Structural basis for an unexpected mode of SERM-mediated ER antagonism. Mol. Cell 18, 413–424 (2005).
Lai, A. C. & Crews, C. M. Induced protein degradation: an emerging drug discovery paradigm. Nat. Rev. Drug Discov. 16, 101–114 (2017).
Naito, M., Ohoka, N., Shibata, N. & Tsukumo, Y. Targeted protein degradation by chimeric small molecules, PROTACs and SNIPERs. Front. Chem. 7, 849 (2019).
Neklesa, T. K. et al. Small-molecule hydrophobic tagging-induced degradation of HaloTag fusion proteins. Nat. Chem. Biol. 7, 538–543 (2011).
Słabicki, M. et al. The CDK inhibitor CR8 acts as a molecular glue degrader that depletes cyclin K. Nature 585, 293–297 (2020).
Ito, T. et al. Identification of a primary target of thalidomide teratogenicity. Science 327, 1345–1350 (2010).
Fischer, E. S. et al. Structure of the DDB1-CRBN E3 ubiquitin ligase in complex with thalidomide. Nature 512, 49–53 (2014).
Krönke, J. et al. Lenalidomide causes selective degradation of IKZF1 and IKZF3 in multiple myeloma cells. Science 343, 301–305 (2014).
Lu, G. et al. The myeloma drug lenalidomide promotes the cereblon-dependent destruction of Ikaros proteins. Science 343, 305–309 (2014).
Krönke, J. et al. Lenalidomide induces ubiquitination and degradation of CK1α in del(5q) MDS. Nature 523, 183–188 (2015).
Banik, S. M. et al. Lysosome-targeting chimeras for degradation of extraceullar proteins. Nature 584, 291–297 (2020).
Takahashi, D. et al. AUTACs: cargo-specific degraders using selective autophagy. Mol. Cell 76, 797–810.e10 (2019).
Li, Z., Zhu, C., Ding, Y., Fei, Y. & Lu, B. ATTEC: a potential new approach to target proteinopathies. Autophagy 16, 185–187 (2020).
Nabet, B. et al. The dTAG system for immediate and target-specific protein degradation. Nat. Chem. Biol. 14, 431–441 (2018).
Fauman, E. B., Rai, B. K. & Huang, E. S. Structure-based druggability assessment — identifying suitable targets for small molecule therapeutics. Curr. Opin. Chem. Biol. 15, 463–468 (2011).
Griffith, M. et al. DGIdb: mining the druggable genome. Nat. Methods 10, 1209–1210 (2013).
Kumar, R. D., Chang, L.-W., Ellis, M. J. & Bose, R. Prioritizing potentially druggable mutations with dGene: an annotation tool for cancer genome sequencing data. PLoS ONE 8, e67980 (2013).
Wang, X., Wang, R., Zhang, Y. & Zhang, H. Evolutionary survey of druggable protein targets with respect to their subcellular localizations. Genome Biol. Evol. 5, 1291–1297 (2013).
Finan, C. et al. The druggable genome and support for target identification and validation in drug development. Sci. Transl Med. 9, eaag1166 (2017).
Rodgers, G. et al. Glimmers in illuminating the druggable genome. Nat. Rev. Drug Discov. 17, 301–302 (2018).
Wang, J., Yazdani, S., Han, A. & Schapira, M. Structure-based view of the druggable genome. Drug Discov. Today 25, 561–567 (2020).
Brown, K. K. et al. Approaches to target tractability assessment – a practical perspective. MedChemComm 9, 606–613 (2018).
Oprea, T. I. et al. Unexplored therapeutic opportunities in the human genome. Nat. Rev. Drug Discov. 17, 317–332 (2018).
Carvalho-Silva, D. et al. Open Targets Platform: new developments and updates two years on. Nucleic Acids Res. 47, D1056–D1065 (2019).
Ochoa, D. et al. Open Targets Platform: supporting systematic drug-target identification and prioritisation. Nucleic Acids Res. 49, D1302–D1310 (2021).
Mullard, A. Targeted protein degraders crowd into the clinic. Nat. Rev. Drug Discov. 20, 247–250 (2021).
Weng, G. et al. PROTAC-DB: an online database of PROTACs. Nucleic Acids Res. 49, D1381–D1387 (2021).
Yang, J. et al. Covalent modification of Cys-239 in β-tubulin by small molecules as a strategy to promote tubulin heterodimer degradation. J. Biol. Chem. 294, 8161–8170 (2019).
Gasic, I. et al. Tubulin resists degradation by cereblon-recruiting PROTACs. Cells 9, 1083 (2020).
Donovan, K. A. et al. Mapping the degradable kinome provides a resource for expedited degrader development. Cell 183, 1714–1731.e10 (2020).
Arrowsmith, C. H. et al. The promise and peril of chemical probes. Nat. Chem. Biol. 11, 536–541 (2015).
UniProt Consortium. UniProt: a worldwide hub of protein knowledge. Nucleic Acids Res. 47, D506–D515 (2019).
Hornbeck, P. V. et al. 15 years of PhosphoSitePlus®: integrating post-translationally modified sites, disease variants and isoforms. Nucleic Acids Res. 47, D433–D441 (2019).
Chen, T. et al. mUbiSiDa: a comprehensive database for protein ubiquitination sites in mammals. PLoS ONE 9, e85744 (2014).
Kim, W. et al. Systematic and quantitative assessment of the ubiquitin-modified proteome. Mol. Cell 44, 325–340 (2011).
Mathieson, T. et al. Systematic analysis of protein turnover in primary cells. Nat. Commun. 9, 689 (2018).
Mendez, D. et al. ChEMBL: towards direct deposition of bioassay data. Nucleic Acids Res. 47, D930–D940 (2019).
Bondeson, D. P. et al. Lessons in PROTAC design from selective degradation with a promiscuous warhead. Cell Chem. Biol. 25, 78–87.e5 (2018).
Testa, A. et al. 3-Fluoro-4-hydroxyprolines: synthesis, conformational analysis, and stereoselective recognition by the VHL E3 ubiquitin ligase for targeted protein degradation. J. Am. Chem. Soc. 140, 9299–9313 (2018).
Han, X. et al. Discovery of highly potent and efficient PROTAC degraders of androgen receptor (AR) by employing weak binding affinity VHL E3 ligase ligands. J. Med. Chem. 62, 11218–11231 (2019).
Bassi, Z. I. et al. Modulating PCAF/GCN5 immune cell function through a PROTAC approach. ACS Chem. Biol. 13, 2862–2867 (2018).
Gechijian, L. N. et al. Functional TRIM24 degrader via conjugation of ineffectual bromodomain and VHL ligands. Nat. Chem. Biol. 14, 405–412 (2018).
Farnaby, W. et al. BAF complex vulnerabilities in cancer demonstrated via structure-based PROTAC design. Nat. Chem. Biol. 15, 672–680 (2019).
The Gene Ontology Consortium. The gene ontology resource: 20 years and still going strong. Nucleic Acids Res. 47, D330–D338 (2019).
Lin, L., Yee, S. W., Kim, R. B. & Giacomini, K. M. SLC transporters as therapeutic targets: emerging opportunities. Nat. Rev. Drug Discov. 14, 543–560 (2015).
Wang, W. W., Gallo, L., Jadhav, A., Hawkins, R. & Parker, C. G. The druggability of solute carriers. J. Med. Chem. 63, 3834–3867 (2020).
Bensimon, A. et al. Targeted degradation of SLC transporters reveals amenability of multi-pass transmembrane proteins to ligand-induced proteolysis. Cell Chem. Biol. 27, 728–739.e9 (2020).
Koscielny, G. et al. Open Targets: a platform for therapeutic target identification and validation. Nucleic Acids Res. 45, D985–D994 (2017).
Christiano, R. et al. A systematic protein turnover map for decoding protein degradation. Cell Rep. 33, 108378 (2020).
Komander, D. & Rape, M. The ubiquitin code. Annu. Rev. Biochem. 81, 203–229 (2012).
Yau, R. & Rape, M. The increasing complexity of the ubiquitin code. Nat. Cell Biol. 18, 579–586 (2016).
Kwon, Y. T. & Ciechanover, A. The ubiquitin code in the ubiquitin-proteasome system and autophagy. Trends Biochem. Sci. 42, 873–886 (2017).
Mattern, M., Sutherland, J., Kadimisetty, K., Barrio, R. & Rodriguez, M. S. Using ubiquitin binders to decipher the ubiquitin code. Trends Biochem. Sci. 44, 599–615 (2019).
Acknowledgements
The authors thank A. Ciulli, E. Fischer and M. Calabrese for their constructive feedback on our original manuscript. This work was funded by the Open Targets consortium and the Member States of the European Molecular Biology Laboratory (EMBL).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
G.H., S.R., V.S., M.M.H., P.J.T., M.A.Q., A.B.B. and K.B. work for companies involved in the discovery of PROTAC compounds as part of their overall drug discovery efforts. The other authors declare no competing interests.
Additional information
Peer review information
Nature Reviews Drug Discovery thanks A. Ciulli, E. Fischer and M. Calabrese for their contribution to the peer review of this work.
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Related Links
Open Targets Platform: https://platform.opentargets.org/
Open Targets scoring scheme: https://platform-docs.opentargets.org/associations
Pharos API: https://pharos.nih.gov/api
PROTACpedia: http://protacdb.weizmann.ac.il/ptcb/main
Supplementary information
Rights and permissions
About this article
Cite this article
Schneider, M., Radoux, C.J., Hercules, A. et al. The PROTACtable genome. Nat Rev Drug Discov 20, 789–797 (2021). https://doi.org/10.1038/s41573-021-00245-x
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41573-021-00245-x
This article is cited by
-
From bench to bedside: current development and emerging trend of KRAS-targeted therapy
Acta Pharmacologica Sinica (2024)
-
PROTAC’ing oncoproteins: targeted protein degradation for cancer therapy
Molecular Cancer (2023)
-
Late-stage synthesis of heterobifunctional molecules for PROTAC applications via ruthenium-catalysed C‒H amidation
Nature Communications (2023)
-
Lenalidomide derivatives and proteolysis-targeting chimeras for controlling neosubstrate degradation
Nature Communications (2023)
-
ALK-positive lung cancer: a moving target
Nature Cancer (2023)