Abstract
Oncogenes that occur in ≤5% of non-small-cell lung cancers have been defined as ‘rare’; nonetheless, this frequency can correspond to a substantial number of patients diagnosed annually. Within rare oncogenes, less commonly identified alterations (such as HRAS, NRAS, RIT1, ARAF, RAF1 and MAP2K1 mutations, or ERBB family, LTK and RASGRF1 fusions) can share certain structural or oncogenic features with more commonly recognized alterations (such as KRAS, BRAF, MET and ERBB family mutations, or ALK, RET and ROS1 fusions). Over the past 5 years, a surge in the identification of rare-oncogene-driven lung cancers has challenged the boundaries of traditional clinical grade diagnostic assays and profiling algorithms. In tandem, the number of approved targeted therapies for patients with rare molecular subtypes of lung cancer has risen dramatically. Rational drug design has iteratively improved the quality of small-molecule therapeutic agents and introduced a wave of antibody-based therapeutics, expanding the list of actionable de novo and resistance alterations in lung cancer. Getting additional molecularly tailored therapeutics approved for rare-oncogene-driven lung cancers in a larger range of countries will require ongoing stakeholder cooperation. Patient advocates, health-care agencies, investigators and companies with an interest in diagnostics, therapeutics and real-world evidence have already taken steps to surmount the challenges associated with research into low-frequency drivers.
Key points
-
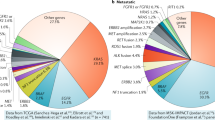
Many ‘rare’ molecular subtypes of lung cancer individually account for a substantial number of patients diagnosed annually around the world.
-
An incredible diversity of molecular subtypes of lung cancer exists; mechanistically, these can be classified into mutations, fusions and copy number changes.
-
Alterations involving receptor tyrosine kinases and MAPK pathway members can share structural and/or oncogenic features; conversely, other alterations have distinct mechanisms of oncogenesis such as effects on RNA splicing or epigenetic processes.
-
Optimizing the identification of rare driver oncogenes requires both clinicopathological approaches that are feature-agnostic and tailored approaches to patient selection, tumour and plasma interrogation, DNA and RNA sequencing, and more unbiased profiling.
-
Targeted therapy approvals were previously focused on certain alterations to the point of saturation and dominated by small-molecule tyrosine kinase inhibitors; approved and investigational antibody-based therapies are now becoming more widely used.
-
Oncogene-driven advocacy, the adoption of contemporary trial designs, expedited regulatory pathways for drug development, and real-world evidence generation are all crucial steps towards promoting research and expediting the approval of drugs for rare oncogene-driven lung cancers.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
References
Tambuyzer, E. et al. Therapies for rare diseases: therapeutic modalities, progress and challenges ahead. Nat. Rev. Drug Discov. 19, 93–111 (2020).
Gatta, G. et al. Rare cancers are not so rare: the rare cancer burden in Europe. Eur. J. Cancer 47, 2493–2511 (2011).
Matsuda, T. et al. Rare cancers are not rare in Asia as well: the rare cancer burden in East Asia. Cancer Epidemiol. 67, 101702 (2020).
National Cancer Institute. NCI Dictionary of Cancer Terms https://www.cancer.gov/publications/dictionaries/cancer-terms/def/rare-cancer (2022).
McCoach, C. E. & Doebele, R. C. The minority report: targeting the rare oncogenes in NSCLC. Curr. Treat. Options Oncol. 15, 644–657 (2014).
Centers for Disease Control and Prevention. Annual trends in new cancers. https://gis.cdc.gov/Cancer/USCS/#/Trends/ (2022).
Guo, Y. et al. Recent progress in rare oncogenic drivers and targeted therapy for non-small cell lung cancer. Onco Targets Ther. 12, 10343–10360 (2019).
Cancer Genome Atlas Research Network. Comprehensive molecular profiling of lung adenocarcinoma. Nature 511, 543–550 (2014).
Siegel, R. L., Miller, K. D., Fuchs, H. E. & Jemal, A. Cancer statistics, 2022. CA Cancer J. Clin. 72, 7–33 (2022).
Tong, J. H. et al. MET amplification and exon 14 splice site mutation define unique molecular subgroups of non-small cell lung carcinoma with poor prognosis. Clin. Cancer Res. 22, 3048–3056 (2016).
Friedlaender, A. et al. EGFR and HER2 exon 20 insertions in solid tumours: from biology to treatment. Nat. Rev. Clin. Oncol. 19, 51–69 (2022).
Hammerman, P. S. et al. Mutations in the DDR2 kinase gene identify a novel therapeutic target in squamous cell lung cancer. Cancer Discov. 1, 78–89 (2011).
The AACR Project GENIE Consortium. AACR Project GENIE: powering precision medicine through an international consortium. Cancer Discov. 7, 818–831 (2017).
Luo, J. et al. Overcoming KRAS-mutant lung cancer. Am. Soc. Clin. Oncol. Educ. Book 42, 1–11 (2022).
Du, Z. et al. Structure–function analysis of oncogenic EGFR kinase domain duplication reveals insights into activation and a potential approach for therapeutic targeting. Nat. Commun. 12, 1382 (2021).
Hunihan, L. et al. RASGRF1 fusions activate oncogenic RAS signaling and confer sensitivity to MEK inhibition. Clin. Cancer Res. 28, 3091–3103 (2022).
Reddy, V. P. et al. BRAF fusions in clinically advanced non-small cell lung cancer: an emerging target for anti-BRAF therapies [abstract]. J. Clin. Oncol. 35 (Suppl. 15), 9072 (2017).
Nagasaka, M. & Ou, S. I. NRG1 and NRG2 fusion positive solid tumor malignancies: a paradigm of ligand-fusion oncogenesis. Trends Cancer 8, 242–258 (2022).
Ou, S. I. et al. Identification of novel CDH1-NRG2α and F11R-NRG2α fusions in NSCLC plus additional novel NRG2α fusions in other solid tumors by whole transcriptome sequencing. JTO Clin. Res. Rep. 2, 100132 (2021).
Costa, F. A. et al. Revealing the BRD4-NOTCH3 fusion: a novel hill in the cancer landscape. Lung Cancer 154, 146–150 (2021).
Van, A. N. et al. Protein kinase C fusion proteins are paradoxically loss of function in cancer. J. Biol. Chem. 296, 100445 (2021).
Liu, S. et al. The genomic characteristics of ALK fusion positive tumors in Chinese NSCLC patients. Front. Oncol. 10, 726 (2020).
Schram, A. M., Chang, M. T., Jonsson, P. & Drilon, A. Fusions in solid tumours: diagnostic strategies, targeted therapy, and acquired resistance. Nat. Rev. Clin. Oncol. 14, 735–748 (2017).
Hechtman, J. F. et al. Pan-Trk immunohistochemistry is an efficient and reliable screen for the detection of NTRK fusions. Am. J. Surg. Pathol. 41, 1547–1551 (2017).
Wang, K. et al. FGFR1-ERK1/2-SOX2 axis promotes cell proliferation, epithelial-mesenchymal transition, and metastasis in FGFR1-amplified lung cancer. Oncogene 37, 5340–5354 (2018).
Su, W. et al. ARAF protein kinase activates RAS by antagonizing its binding to RASGAP NF1. Mol. Cell 82, 2443–2457.e7 (2022).
Kim, H. et al. Extrachromosomal DNA is associated with oncogene amplification and poor outcome across multiple cancers. Nat. Genet. 52, 891–897 (2020).
Yu, H. A. et al. Analysis of tumor specimens at the time of acquired resistance to EGFR-TKI therapy in 155 patients with EGFR-mutant lung cancers. Clin. Cancer Res. 19, 2240–2247 (2013).
Zhao, J. & Xia, Y. Targeting HER2 alterations in non-small-cell lung cancer: a comprehensive review. JCO Precis. Oncol. 4, 411–425 (2020).
Zabransky, D. J. et al. HER2 missense mutations have distinct effects on oncogenic signaling and migration. Proc. Natl Acad. Sci. USA 112, E6205–E6214 (2015).
Vyse, S. & Huang, P. H. Targeting EGFR exon 20 insertion mutations in non-small cell lung cancer. Signal. Transduct. Target. Ther. 4, 5 (2019).
Neel, D. S. et al. Differential subcellular localization regulates oncogenic signaling by ROS1 kinase fusion proteins. Cancer Res. 79, 546–556 (2019).
Guo, R. et al. MET-dependent solid tumours – molecular diagnosis and targeted therapy. Nat. Rev. Clin. Oncol. 17, 569–587 (2020).
Zingg, D. et al. Truncated FGFR2 is a clinically actionable oncogene in multiple cancers. Nature 608, 609–617 (2022).
Smith, H. W. et al. An ErbB2 splice variant lacking exon 16 drives lung carcinoma. Proc. Natl Acad. Sci. USA 117, 20139–20148 (2020).
Haigis, K. M. KRAS alleles: the devil is in the detail. Trends Cancer 3, 686–697 (2017).
Hayashi, T. et al. RASA1 and NF1 are preferentially co-mutated and define a distinct genetic subset of smoking-associated non-small cell lung carcinomas sensitive to MEK inhibition. Clin. Cancer Res. 24, 1436–1447 (2018).
Castel, P. et al. RIT1 oncoproteins escape LZTR1-mediated proteolysis. Science 363, 1226–1230 (2019).
Yao, Z. et al. Tumours with class 3 BRAF mutants are sensitive to the inhibition of activated RAS. Nature 548, 234–238 (2017).
Noeparast, A. et al. CRAF mutations in lung cancer can be oncogenic and predict sensitivity to combined type II RAF and MEK inhibition. Oncogene 38, 5933–5941 (2019).
Arcila, M. E. et al. MAP2K1 (MEK1) mutations define a distinct subset of lung adenocarcinoma associated with smoking. Clin. Cancer Res. 21, 1935–1943 (2015).
French, C. NUT midline carcinoma. Nat. Rev. Cancer 14, 149–150 (2014).
Wang, K. et al. PEST domain mutations in Notch receptors comprise an oncogenic driver segment in triple-negative breast cancer sensitive to a gamma-secretase inhibitor. Clin. Cancer Res. 21, 1487–1496 (2015).
Jordan, E. J. et al. Prospective comprehensive molecular characterization of lung adenocarcinomas for efficient patient matching to approved and emerging therapies. Cancer Discov. 7, 596–609 (2017).
Perez-Moreno, P., Brambilla, E., Thomas, R. & Soria, J. C. Squamous cell carcinoma of the lung: molecular subtypes and therapeutic opportunities. Clin. Cancer Res. 18, 2443–2451 (2012).
Nishino, M. et al. Histologic and cytomorphologic features of ALK-rearranged lung adenocarcinomas. Mod. Pathol. 25, 1462–1472 (2012).
Lee, S. E. et al. Comprehensive analysis of RET and ROS1 rearrangement in lung adenocarcinoma. Mod. Pathol. 28, 468–479 (2015).
Nagasaka, M. & Ou, S. I. Neuregulin 1 fusion-positive NSCLC. J. Thorac. Oncol. 14, 1354–1359 (2019).
Chang, J. C. et al. Comprehensive molecular and clinicopathologic analysis of 200 pulmonary invasive mucinous adenocarcinomas identifies distinct characteristics of molecular subtypes. Clin. Cancer Res. 27, 4066–4076 (2021).
Shim, H. S. et al. Unique genetic and survival characteristics of invasive mucinous adenocarcinoma of the lung. J. Thorac. Oncol. 10, 1156–1162 (2015).
Lee, J. K. et al. Characterization of non-small-cell lung cancers with MET exon 14 skipping alterations detected in tissue or liquid: clinicogenomics and real-world treatment patterns. JCO Precis. Oncol. https://doi.org/10.1200/PO.21.00122 (2021).
Socinski, M. A., Pennell, N. A. & Davies, K. D. MET exon 14 skipping mutations in non-small-cell lung cancer: an overview of biology, clinical outcomes, and testing considerations. JCO Precis. Oncol. https://doi.org/10.1200/PO.20.00516 (2021).
Al-Samkari, H. et al. Impact of ALK rearrangement on venous and arterial thrombotic risk in NSCLC. J. Thorac. Oncol. 15, 1497–1506 (2020).
Ng, T. L. et al. ROS1 gene rearrangements are associated with an elevated risk of peridiagnosis thromboembolic events. J. Thorac. Oncol. 14, 596–605 (2019).
Shaw, A. T. & Engelman, J. A. ALK in lung cancer: past, present, and future. J. Clin. Oncol. 31, 1105–1111 (2013).
Tsuta, K. et al. RET-rearranged non-small-cell lung carcinoma: a clinicopathological and molecular analysis. Br. J. Cancer 110, 1571–1578 (2014).
Parikh, D. A. et al. Characteristics of patients with ROS1+ cancers: results from the first patient-designed, global, pan-cancer ROS1 data repository. JCO Oncol. Pract. 16, e183–e189 (2020).
Awad, M. M. et al. MET exon 14 mutations in non-small-cell lung cancer are associated with advanced age and stage-dependent MET genomic amplification and c-Met overexpression. J. Clin. Oncol. 34, 721–730 (2016).
Riely, G. J. et al. Frequency and distinctive spectrum of KRAS mutations in never smokers with lung adenocarcinoma. Clin. Cancer Res. 14, 5731–5734 (2008).
Camidge, D. R. et al. Anaplastic lymphoma kinase gene rearrangements in non-small cell lung cancer are associated with prolonged progression-free survival on pemetrexed. J. Thorac. Oncol. 6, 774–780 (2011).
Drilon, A. et al. Clinical outcomes with pemetrexed-based systemic therapies in RET-rearranged lung cancers. Ann. Oncol. 27, 1286–1291 (2016).
Chen, Y. F. et al. Efficacy of pemetrexed-based chemotherapy in patients with ROS1 fusion-positive lung adenocarcinoma compared with in patients harboring other driver mutations in East Asian populations. J. Thorac. Oncol. 11, 1140–1152 (2016).
Marchetti, A. et al. Clinical features and outcome of patients with non-small-cell lung cancer harboring BRAF mutations. J. Clin. Oncol. 29, 3574–3579 (2011).
Wang, Y. et al. Outcomes of pemetrexed-based chemotherapies in HER2-mutant lung cancers. BMC Cancer 18, 326 (2018).
Naidoo, J. et al. Epidermal growth factor receptor exon 20 insertions in advanced lung adenocarcinomas: clinical outcomes and response to erlotinib. Cancer 121, 3212–3220 (2015).
Rosen, E. Y. et al. TRK fusions are enriched in cancers with uncommon histologies and the absence of canonical driver mutations. Clin. Cancer Res. 26, 1624–1632 (2020).
Drilon, A. et al. Clinicopathologic features and response to therapy of NRG1 fusion-driven lung cancers: the eNRGy1 global multicenter registry. J. Clin. Oncol. 39, 2791–2802 (2021).
Mazieres, J. et al. Immune checkpoint inhibitors for patients with advanced lung cancer and oncogenic driver alterations: results from the IMMUNOTARGET registry. Ann. Oncol. 30, 1321–1328 (2019).
Negrao, M. V. et al. Oncogene-specific differences in tumor mutational burden, PD-L1 expression, and outcomes from immunotherapy in non-small cell lung cancer. J. Immunother. Cancer https://doi.org/10.1136/jitc-2021-002891 (2021).
Guisier, F. et al. Brief report: First-line pembrolizumab in metastatic non-small cell lung cancer habouring MET exon 14 skipping mutation and PD-L1 ≥50% (GFPC 01-20 study). Clin. Lung Cancer 23, e545–e549 (2022).
Schoenfeld, A. J. et al. Severe immune-related adverse events are common with sequential PD-(L)1 blockade and osimertinib. Ann. Oncol. 30, 839–844 (2019).
Lin, J. J. et al. Increased hepatotoxicity associated with sequential immune checkpoint inhibitor and crizotinib therapy in patients with non-small cell lung cancer. J. Thorac. Oncol. 14, 135–140 (2019).
McCoach, C. E. et al. Hypersensitivity reactions to selpercatinib treatment with or without prior immune checkpoint inhibitor therapy in patients with NSCLC in LIBRETTO-001. J. Thorac. Oncol. 17, 768–778 (2022).
Yang, S. R. et al. Precision medicine in non-small cell lung cancer: current applications and future directions. Semin. Cancer Biol. 84, 184–198 (2022).
Lindeman, N. I. et al. Molecular testing guideline for selection of lung cancer patients for EGFR and ALK tyrosine kinase inhibitors: guideline from the College of American Pathologists, International Association for the Study of Lung Cancer, and Association for Molecular Pathology. J. Thorac. Oncol. 8, 823–859 (2013).
Lindeman, N. I. et al. Updated molecular testing guideline for the selection of lung cancer patients for treatment with targeted tyrosine kinase inhibitors: guideline from the College of American Pathologists, the International Association for the Study of Lung Cancer, and the Association for Molecular Pathology. Arch. Pathol. Lab. Med. 142, 321–346 (2018).
Cheng, D. T. et al. Memorial Sloan Kettering-integrated mutation profiling of actionable cancer targets (MSK-IMPACT): a hybridization capture-based next-generation sequencing clinical assay for solid tumor molecular oncology. J. Mol. Diagn. 17, 251–264 (2015).
Passiglia, F., Malapelle, U., Normanno, N. & Pinto, C. Optimizing diagnosis and treatment of EGFR exon 20 insertions mutant NSCLC. Cancer Treat. Rev. 109, 102438 (2022).
Drilon, A., Cappuzzo, F., Ou, S. I. & Camidge, D. R. Targeting MET in lung cancer: will expectations finally be MET? J. Thorac. Oncol. 12, 15–26 (2017).
Davies, K. D. et al. Comparison of molecular testing modalities for detection of ROS1 rearrangements in a cohort of positive patient samples. J. Thorac. Oncol. 13, 1474–1482 (2018).
Benayed, R. et al. High yield of RNA sequencing for targetable kinase fusions in lung adenocarcinomas with no mitogenic driver alteration detected by DNA sequencing and low tumor mutation burden. Clin. Cancer Res. 25, 4712–4722 (2019).
Drilon, A. et al. Response to ERBB3-directed targeted therapy in NRG1-rearranged cancers. Cancer Discov. 8, 686–695 (2018).
Yang, S. R. et al. A performance comparison of commonly used assays to detect RET fusions. Clin. Cancer Res. 27, 1316–1328 (2021).
Poirot, B. et al. MET exon 14 alterations and new resistance mutations to tyrosine kinase inhibitors: risk of inadequate detection with current amplicon-based NGS panels. J. Thorac. Oncol. 12, 1582–1587 (2017).
Davies, K. D. et al. DNA-based versus RNA-based detection of MET exon 14 skipping events in lung cancer. J. Thorac. Oncol. 14, 737–741 (2019).
Guo, R. et al. MET exon 14-altered lung cancers and MET inhibitor resistance. Clin. Cancer Res. 27, 799–806 (2021).
Desmeules, P. et al. Performance of an RNA-based next-generation sequencing assay for combined detection of clinically actionable fusions and hotspot mutations in NSCLC. JTO Clin. Res. Rep. 3, 100276 (2022).
Cohen, D. et al. Optimizing mutation and fusion detection in NSCLC by sequential DNA and RNA sequencing. J. Thorac. Oncol. 15, 1000–1014 (2020).
Hovelson, D. H. et al. Development and validation of a scalable next-generation sequencing system for assessing relevant somatic variants in solid tumors. Neoplasia 17, 385–399 (2015).
Solomon, J. P. et al. Bioinformatically-expanded next-generation sequencing analysis optimizes identification of therapeutically relevant MET copy number alterations in >50,000 tumors. Clin. Cancer Res. https://doi.org/10.1158/1078-0432.CCR-22-1321 (2022).
Schrock, A. B. et al. Characterization of 298 patients with lung cancer harboring MET exon 14 skipping alterations. J. Thorac. Oncol. 11, 1493–1502 (2016).
Wolf, J. et al. Capmatinib in MET exon 14-mutated or MET-amplified non-small-cell lung cancer. N. Engl. J. Med. 383, 944–957 (2020).
Al-Kateb, H., Nguyen, T. T., Steger-May, K. & Pfeifer, J. D. Identification of major factors associated with failed clinical molecular oncology testing performed by next generation sequencing (NGS). Mol. Oncol. 9, 1737–1743 (2015).
Leighl, N. B. et al. Clinical utility of comprehensive cell-free DNA analysis to identify genomic biomarkers in patients with newly diagnosed metastatic non-small cell lung cancer. Clin. Cancer Res. 25, 4691–4700 (2019).
Brannon, A. R. et al. Enhanced specificity of clinical high-sensitivity tumor mutation profiling in cell-free DNA via paired normal sequencing using MSK-ACCESS. Nat. Commun. 12, 3770 (2021).
Finkle, J. D. et al. Validation of a liquid biopsy assay with molecular and clinical profiling of circulating tumor DNA. NPJ Precis. Oncol. 5, 63 (2021).
Woodhouse, R. et al. Clinical and analytical validation of FoundationOne Liquid CDx, a novel 324-gene cfDNA-based comprehensive genomic profiling assay for cancers of solid tumor origin. PLoS ONE 15, e0237802 (2020).
Bale, T. A. et al. Clinical experience of cerebrospinal fluid-based liquid biopsy demonstrates superiority of cell-free DNA over cell pellet genomic DNA for molecular profiling. J. Mol. Diagn. 23, 742–752 (2021).
Yang, S. R. et al. Targeted deep sequencing of cell-free DNA in serous body cavity fluids with malignant, suspicious, and benign cytology. Cancer Cytopathol. 128, 43–56 (2020).
Negishi, R. et al. Transcriptomic profiling of single circulating tumor cells provides insight into human metastatic gastric cancer. Commun. Biol. 5, 20 (2022).
Hasegawa, N. et al. Highly sensitive fusion detection using plasma cell-free RNA in non-small-cell lung cancers. Cancer Sci. 112, 4393–4403 (2021).
Dunwell, T. L. et al. Adaptor template oligo-mediated sequencing (ATOM-Seq) is a new ultra-sensitive UMI-based NGS library preparation technology for use with cfDNA and cfRNA. Sci. Rep. 11, 3138 (2021).
Wang, M. C. et al. Methods for collection of extracellular vesicles and their content RNA as liquid biopsy for lung cancer detection: application of differential centrifugation and annexin A5 coated beads. Curr. Issues Mol. Biol. 44, 2374–2386 (2022).
Izumi, H. et al. The CLIP1-LTK fusion is an oncogenic driver in non-small-cell lung cancer. Nature 600, 319–323 (2021).
Carrot-Zhang, J. et al. Whole-genome characterization of lung adenocarcinomas lacking the RTK/RAS/RAF pathway. Cell Rep. 34, 108707 (2021).
Roskoski, R. Jr Properties of FDA-approved small molecule protein kinase inhibitors: a 2022 update. Pharmacol. Res. 175, 106037 (2022).
Dar, A. C. & Shokat, K. M. The evolution of protein kinase inhibitors from antagonists to agonists of cellular signaling. Annu. Rev. Biochem. 80, 769–795 (2011).
Yao, Z. et al. RAF inhibitor PLX8394 selectively disrupts BRAF dimers and RAS-independent BRAF-mutant-driven signaling. Nat. Med. 25, 284–291 (2019).
Alabi, S. et al. Mutant-selective degradation by BRAF-targeting PROTACs. Nat. Commun. 12, 920 (2021).
Li, J. W., Zheng, G., Kaye, F. J. & Wu, L. PROTAC therapy as a new targeted therapy for lung cancer. Mol. Ther. https://doi.org/10.1016/j.ymthe.2022.11.011 (2022).
Burslem, G. M. et al. The advantages of targeted protein degradation over inhibition: an RTK case study. Cell Chem. Biol. 25, 67–77.e3 (2018).
Kinoshita, I. et al. A phase II study of trastuzumab monotherapy in pretreated patients with non-small cell lung cancers (NSCLCs) harboring HER2 alterations: HOT1303-B trial. Ann. Oncol. 29, viii540 (2018).
Drago, J. Z., Modi, S. & Chandarlapaty, S. Unlocking the potential of antibody–drug conjugates for cancer therapy. Nat. Rev. Clin. Oncol. 18, 327–344 (2021).
Ackerman, S. E. et al. Immune-stimulating antibody conjugates elicit robust myeloid activation and durable antitumor immunity. Nat. Cancer 2, 18–33 (2021).
Li, B. T. et al. Trastuzumab deruxtecan in HER2-mutant non-small-cell lung cancer. N. Engl. J. Med. 386, 241–251 (2021).
Camidge, D. R. et al. Telisotuzumab vedotin (Teliso-V) monotherapy in patients (pts) with previously treated c-Met–overexpressing (OE) advanced non-small cell lung cancer (NSCLC) [abstract]. J. Clin. Oncol. 40 (Suppl. 16), 9016 (2022).
Paik, P. K. et al. Tepotinib in non-small-cell lung cancer with MET exon 14 skipping mutations. N. Engl. J. Med. 383, 931–943 (2020).
Planchard, D. et al. Dabrafenib plus trametinib in patients with previously untreated BRAFV600E-mutant metastatic non-small-cell lung cancer: an open-label, phase 2 trial. Lancet Oncol. 18, 1307–1316 (2017).
Zhou, C. et al. Treatment outcomes and safety of mobocertinib in platinum-pretreated patients with EGFR exon 20 insertion-positive metastatic non-small cell lung cancer: a phase 1/2 open-label nonrandomized clinical trial. JAMA Oncol. 7, e214761 (2021).
Park, K. et al. Amivantamab in EGFR exon 20 insertion-mutated non-small-cell lung cancer progressing on platinum chemotherapy: initial results from the CHRYSALIS phase I study. J. Clin. Oncol. 39, 3391–3402 (2021).
Krebs, M. et al. Amivantamab in patients with NSCLC with MET exon 14 skipping mutation: updated results from the CHRYSALIS study [abstract]. J. Clin. Oncol. 40 (Suppl. 16), 9008 (2022).
Zhou, C. et al. Pyrotinib in HER2-mutant advanced lung adenocarcinoma after platinum-based chemotherapy: a multicenter, open-label, single-arm, phase II study. J. Clin. Oncol. 38, 2753–2761 (2020).
Elamin, Y. Y. et al. Poziotinib for patients with HER2 exon 20 mutant non-small-cell lung cancer: results from a phase II trial. J. Clin. Oncol. 40, 702–709 (2022).
Li, B. T. et al. Ado-trastuzumab emtansine for patients with HER2-mutant lung cancers: results from a phase II basket trial. J. Clin. Oncol. 36, 2532–2537 (2018).
Mao, Z. et al. KRAS(G12D) can be targeted by potent inhibitors via formation of salt bridge. Cell Discov. 8, 5 (2022).
Planchard, D. et al. Dabrafenib in patients with BRAF(V600E)-positive advanced non-small-cell lung cancer: a single-arm, multicentre, open-label, phase 2 trial. Lancet Oncol. 17, 642–650 (2016).
Subbiah, V. et al. Efficacy of vemurafenib in patients with non-small-cell lung cancer with BRAF V600 mutation: an open-label, single-arm cohort of the histology-independent VE-BASKET study. JCO Precis. Oncol. 3, PO.18.00266 (2019).
Berger, A. H. et al. Oncogenic RIT1 mutations in lung adenocarcinoma. Oncogene 33, 4418–4423 (2014).
Moreno, V. et al. Extended follow-up of efficacy and safety of larotrectinib in patients with TRK fusion lung cancer [abstract EP08.02-148]. J. Thorac. Oncol. 17 (Suppl. 9), S473–S474 (2022).
Drilon, A. et al. ROS1-dependent cancers – biology, diagnostics and therapeutics. Nat. Rev. Clin. Oncol. 18, 35–55 (2021).
Alexander, M. et al. A multicenter study of thromboembolic events among patients diagnosed with ROS1-rearranged non-small cell lung cancer. Lung Cancer 142, 34–40 (2020).
Shaw, A. T. et al. Crizotinib in ROS1-rearranged advanced non-small-cell lung cancer (NSCLC): updated results, including overall survival, from PROFILE 1001. Ann. Oncol. 30, 1121–1126 (2019).
Drilon, A. et al. Long-term efficacy and safety of entrectinib in ROS1 fusion-positive NSCLC. JTO Clin. Res. Rep. 3, 100332 (2022).
Drilon, A. et al. Efficacy of selpercatinib in RET fusion-positive non-small-cell lung cancer. N. Engl. J. Med. 383, 813–824 (2020).
Gainor, J. F. et al. Pralsetinib for RET fusion-positive non-small-cell lung cancer (ARROW): a multi-cohort, open-label, phase 1/2 study. Lancet Oncol. 22, 959–969 (2021).
Solomon, B. J. et al. First-line crizotinib versus chemotherapy in ALK-positive lung cancer. N. Engl. J. Med. 371, 2167–2177 (2014).
Soria, J.-C. et al. First-line ceritinib versus platinum-based chemotherapy in advanced ALK-rearranged non-small-cell lung cancer (ASCEND-4): a randomised, open-label, phase 3 study. Lancet 389, 917–929 (2017).
Peters, S. et al. Alectinib versus crizotinib in untreated ALK-positive non-small-cell lung cancer. N. Engl. J. Med. 377, 829–838 (2017).
Mok, T. et al. Updated overall survival and final progression-free survival data for patients with treatment-naive advanced ALK-positive non-small-cell lung cancer in the ALEX study. Ann. Oncol. 31, 1056–1064 (2020).
Camidge, D. R. et al. Brigatinib versus crizotinib in ALK-positive non-small-cell lung cancer. N. Engl. J. Med. 379, 2027–2039 (2018).
Shaw, A. T. et al. First-line lorlatinib or crizotinib in advanced ALK-positive lung cancer. N. Engl. J. Med. 383, 2018–2029 (2020).
Shaw, A. T. et al. Lorlatinib in non-small-cell lung cancer with ALK or ROS1 rearrangement: an international, multicentre, open-label, single-arm first-in-man phase 1 trial. Lancet Oncol. 18, 1590–1599 (2017).
Schram, A. M. et al. Efficacy and safety of zenocutuzumab, a HER2 x HER3 bispecific antibody, across advanced NRG1 fusion (NRG1+) cancers [abstract]. J. Clin. Oncol. 40 (Suppl. 16), 105 (2022).
Carrizosa, D. R. et al. CRESTONE: initial efficacy and safety of seribantumab in solid tumors harboring NRG1 fusions [abstract]. J. Clin. Oncol. 40 (Suppl. 16), 3006 (2022).
Janne, P. A. et al. Efficacy and safety of patritumab deruxtecan (HER3-DXd) in EGFR inhibitor-resistant, EGFR-mutated non-small cell lung cancer. Cancer Discov. 12, 74–89 (2022).
Davies, K. D. et al. Dramatic response to crizotinib in a patient with lung cancer positive for an HLA-DRB1-MET gene fusion. JCO Precis. Oncol. https://doi.org/10.1200/PO.17.00117 (2017).
Qin, A. et al. Detection of known and novel FGFR fusions in non-small cell lung cancer by comprehensive genomic profiling. J. Thorac. Oncol. 14, 54–62 (2019).
Cooper, A. J. et al. Identification of a RAS-activating TMEM87A-RASGRF1 fusion in an exceptional responder to sunitinib with non-small cell lung cancer. Clin. Cancer Res. 26, 4072–4079 (2020).
Camidge, D. R. et al. Crizotinib in patients with MET-amplified NSCLC. J. Thorac. Oncol. 16, 1017–1029 (2021).
Le, X. et al. Tepotinib in patients (pts) with advanced non-small cell lung cancer (NSCLC) with MET amplification (METamp) [abstract]. J. Clin. Oncol. 39 (Suppl. 15), 9021 (2021).
Hainsworth, J. D. et al. Targeted therapy for advanced solid tumors on the basis of molecular profiles: results from MyPathway, an open-label, phase IIa multiple basket study. J. Clin. Oncol. 36, 536–542 (2018).
Song, Z. et al. Pyrotinib in patients with HER2-amplified advanced non-small cell lung cancer: a prospective, multicenter, single-arm trial. Clin. Cancer Res. 28, 461–467 (2022).
Drilon, A. et al. Cabozantinib in patients with advanced RET-rearranged non-small-cell lung cancer: an open-label, single-centre, phase 2, single-arm trial. Lancet Oncol. 17, 1653–1660 (2016).
Lee, S. H. et al. Vandetanib in pretreated patients with advanced non-small cell lung cancer-harboring RET rearrangement: a phase II clinical trial. Ann. Oncol. 28, 292–297 (2017).
Yakes, F. M. et al. Cabozantinib (XL184), a novel MET and VEGFR2 inhibitor, simultaneously suppresses metastasis, angiogenesis, and tumor growth. Mol. Cancer Ther. 10, 2298–2308 (2011).
Demetri, G. D. et al. Updated integrated analysis of the efficacy and safety of entrectinib in patients with NTRK fusion-positive solid tumors. Clin. Cancer Res. 28, 1302–1312 (2022).
Yu, H. A. et al. Phase (Ph) 1/2a study of CLN-081 in patients (pts) with NSCLC with EGFR exon 20 insertion mutations (Ins20) [abstract]. J. Clin. Oncol. 40 (Suppl. 16), 9007 (2022).
Drilon, A. et al. Antitumor activity of crizotinib in lung cancers harboring a MET exon 14 alteration. Nat. Med. 26, 47–51 (2020).
Patil, T. et al. The incidence of brain metastases in stage IV ROS1-rearranged non-small cell lung cancer and rate of central nervous system progression on crizotinib. J. Thorac. Oncol. 13, 1717–1726 (2018).
Shaw, A. T. et al. Lorlatinib in advanced ROS1-positive non-small-cell lung cancer: a multicentre, open-label, single-arm, phase 1-2 trial. Lancet Oncol. 20, 1691–1701 (2019).
Bartsch, R. et al. Trastuzumab deruxtecan in HER2-positive breast cancer with brain metastases: a single-arm, phase 2 trial. Nat. Med. 28, 1840–1847 (2022).
Shu, C. A. et al. Amivantamab and lazertinib in patients with EGFR-mutant non-small cell lung (NSCLC) after progression on osimertinib and platinum-based chemotherapy: updated results from CHRYSALIS-2 [abstract]. J. Clin. Oncol. 40 (Suppl. 16), 9006 (2022).
Aragon-Sanabria, V. et al. Ultrasmall nanoparticle delivery of doxorubicin improves therapeutic index for high-grade glioma. Clin. Cancer Res. 28, 2938–2952 (2022).
Cocco, E. et al. TRK xDFG mutations trigger a sensitivity switch from type I to II kinase inhibitors. Cancer Discov. 11, 126–141 (2021).
Murray, B. W. et al. TPX-0131, a potent CNS-penetrant, next-generation inhibitor of wild-type ALK and ALK-resistant mutations. Mol. Cancer Ther. 20, 1499–1507 (2021).
Pelish, H. E. et al. NUV-655 (NVL-655) is a selective, brain-penetrant ALK inhibitor with antitumor activity against the lorlatinib-resistant G1202R/L1196M compound mutation [abstract]. Cancer Res. 81 (Suppl. 13), 1468 (2021).
Yun, M. R. et al. Repotrectinib exhibits potent antitumor activity in treatment-naïve and solvent-front-mutant ROS1-rearranged non-small cell lung cancer. Clin. Cancer Res. 26, 3287–3295 (2020).
Rotow, J. et al. Combination osimertinib plus selpercatinib for EGFR-mutant non-small cell lung cancer (NSCLC) with acquired RET fusions [abstract FP14.07]. J. Thorac. Oncol. 16 (Suppl. 3), S230 (2021).
Dolgin, E. Oncogene-specific advocacy groups bring a patient-centric perspective to studies of lung cancer. Nature 587, S16–S17 (2020).
Global ROS1 Initiative. The ROS1ders https://www.theros1ders.org/global-ros1-initiative (accessed 10 February 2023).
Maemondo, M. et al. Gefitinib or chemotherapy for non-small-cell lung cancer with mutated EGFR. N. Engl. J. Med. 362, 2380–2388 (2010).
Kim, E. S. et al. The BATTLE trial: personalizing therapy for lung cancer. Cancer Discov. 1, 44–53 (2011).
Herbst, R. S. et al. Lung master protocol (Lung-MAP) – a biomarker-driven protocol for accelerating development of therapies for squamous cell lung cancer: SWOG S1400. Clin. Cancer Res. 21, 1514–1524 (2015).
Middleton, G. et al. The National Lung Matrix trial of personalized therapy in lung cancer. Nature 583, 807–812 (2020).
Hyman, D. M., Taylor, B. S. & Baselga, J. Implementing genome-driven oncology. Cell 168, 584–599 (2017).
Farago, A. F. et al. Clinicopathologic features of non-small-cell lung cancer harboring an NTRK gene fusion. JCO Precis. Oncol. https://doi.org/10.1200/po.18.00037 (2018).
Yang, J. C. H. et al. Rationale and design of ON-TRK: a novel prospective non-interventional study in patients with TRK fusion cancer treated with larotrectinib. BMC Cancer 22, 625 (2022).
Hobbs, B. P. et al. Seamless designs: current practice and considerations for early-phase drug development in oncology. J. Natl Cancer Inst. 111, 118–128 (2019).
Bhatt, D. L. & Mehta, C. Adaptive designs for clinical trials. N. Engl. J. Med. 375, 65–74 (2016).
Vokinger, K. N. & Kesselheim, A. S. Application of orphan drug designation to cancer treatments (2008–2017): a comprehensive and comparative analysis of the USA and EU. BMJ Open. 9, e028634 (2019).
US Food & Drug Administration. Fast track, breakthrough therapy, accelerated approval, priority review. FDA https://www.fda.gov/patients/learn-about-drug-and-device-approvals/fast-track-breakthrough-therapy-accelerated-approval-priority-review (2018).
Kloda, J. H. & Somerville, S. FDA’s expedited review process: the need for speed. appliedclinicaltrialsonline.com, https://www.appliedclinicaltrialsonline.com/view/fda-s-expedited-review-process-need-speed (11 March 2015).
Chodankar, D. Introduction to real-world evidence studies. Perspect. Clin. Res. 12, 171–174 (2021).
Gautschi, O. et al. Targeting RET in patients with RET-rearranged lung cancers: results from the global, multicenter RET registry. J. Clin. Oncol. 35, 1403–1410 (2017).
US Food & Drug Administration. Framework for FDA’s real-world evidence program. FDA https://www.fda.gov/media/120060/download (2018).
Elamin, Y. Y. et al. Poziotinib for EGFR exon 20-mutant NSCLC: clinical efficacy resistance mechanisms and impact of insertion location on drug sensitivity. Cancer Cell 40, 754–767.e6 (2022).
Mazieres, J. et al. Combination of trastuzumab, pertuzumab, and docetaxel in patients with advanced non-small-cell lung cancer harboring HER2 mutations: results from the IFCT-1703 R2D2 trial. J. Clin. Oncol. 40, 719–728 (2022).
Huang, X. et al. The efficacy of ado-trastuzumab emtansine in patients with ERBB2-aberrant non-small cell lung cancer: a systematic review. Transl. Cancer Res. 9, 4507–4516 (2020).
Lu, S. et al. Phase II study of savolitinib in patients (pts) with pulmonary sarcomatoid carcinoma (PSC) and other types of non-small cell lung cancer (NSCLC) harboring MET exon 14 skipping mutations (METex14+) [abstract]. J. Clin. Oncol. 38 (Suppl. 15), 9519 (2020).
Horn, L. et al. Ensartinib vs crizotinib for patients with anaplastic lymphoma kinase-positive non-small cell lung cancer: a randomized clinical trial. JAMA Oncol. 7, 1617–1625 (2021).
Lim, S. M. et al. Open-label, multicenter, phase II study of ceritinib in patients with non-small-cell lung cancer harboring ROS1 rearrangement. J. Clin. Oncol. 35, 2613–2618 (2017).
Turning Point Therapeutics. Turning Point Therapeutics announces positive topline data by blinded independent central review for repotrectinib across all ROS1-positive NSCLC cohorts of phase 1/2 TRIDENT-1 study. Turning Point Therapeutics https://www.tptherapeutics.com/news-releases/news-release-details/turning-point-therapeutics-announces-positive-topline-data/ (2022).
Sanchez-Vega, F. et al. Oncogenic signaling pathways in the cancer genome atlas. Cell 173, 321–337.e10 (2018).
Chen, J. et al. Genomic landscape of lung adenocarcinoma in East Asians. Nat. Genet. 52, 177–186 (2020).
Cerami, E. et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2, 401–404 (2012).
Gao, J. et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci. Signal. 6, pl1 (2013).
ASCO.org. Lung cancer - non-small cell: statistics. Cancer.Net https://www.cancer.net/cancer-types/lung-cancer-non-small-cell/statistics (2022).
Chang, M. T. et al. Identifying recurrent mutations in cancer reveals widespread lineage diversity and mutational specificity. Nat. Biotechnol. 34, 155–163 (2016).
Hida, T. et al. A phase 2 study of lenvatinib in patients with RET fusion-positive lung adenocarcinoma. Lung Cancer 138, 124–130 (2019).
Acknowledgements
The authors to acknowledge their senior editor at Memorial Sloan-Kettering Cancer Center, C. Wilhelm, for the exceptional editorial support he provided for this manuscript. This work was supported in part by an NIH award P30 CA008748. E.C. gratefully acknowledges support from the Lung Cancer Research Foundation (LCRF), the Madelon Ravlin Grant Memorial Award from the Woman’s Cancer Association of the University of Miami and the Tumor Biology Intra-Programmatic Pilot Award from the Sylvester Comprehensive Cancer Center. E.C. also thanks the LCRF for the 2022 William C. Rippe Award.
Author information
Authors and Affiliations
Contributions
All authors made a substantial contribution to all aspects of the preparation of this manuscript.
Corresponding authors
Ethics declarations
Competing interests
G.H. has acted as an adviser to AstraZeneca, Bayer, Lilly, Merck and MSD. S.-R.Y. has received honoraria from PRIME Education LLC. E.C. has acted as a consultant of ENTOS Inc., and has received research funding from ERASCA and InnoCare pharma. A.D. has acted as an adviser to Abbvie, AiCME, Amgen, Applied Pharmaceutical Science, ArcherDX, AstraZeneca, AXIS, Beigene, BergenBio, Blueprint Medicines, Chugai Pharmaceutical, EMD Serono, Entos, EPG Health, Equity Treeline Bio, Exelixis, Harborside Nexus, Helsinn, Hengrui Therapeutics, i3 Health, Ignyta/Genentech/Roche, Janssen, Liberum, Loxo/Bayer/Lilly, mBrace, Medendi, Merus, Monopteros, MORE Health, MonteRosa, 14ner/Elevation Oncology, Novartis, Nuvalent, Ology, Pfizer, Prelude Inc., Remedica Ltd, Repare RX, RV More, Takeda/Ariad/Millenium, TouchIME, TP Therapeutics, Treeline Bio, Tyra Biosciences and Verastem; receives royalties from Wolters Kluwer; receives CME honoraria from Axis, Answers in CME, Clinical Care Options, EPG Health, Imedex, JNCC/Harborside, Liberum, Med Learning, Medscape, MJH Life Sciences, OncLive, Paradigm Medical Communications, Peerview Institute, PeerVoice, Physicians Education Resources, Remedica Ltd, Research to Practice, Targeted Oncology & WebMD; receives institutional research funding from Exelixis, GlaxoSmithKlein, Pfizer, PharmaMar, Taiho and Teva; receives other support from Boehringer Ingelheim; Merck, Merus & Puma; and is listed on a copyright application relating to the use of selpercatinib–osimertinib.
Peer review
Peer review information
Nature Reviews Clinical Oncology thanks the anonymous reviewers for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Related link
Biomarker Collaborative: https://biomarkercollaborative.org/
Supplementary information
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Harada, G., Yang, SR., Cocco, E. et al. Rare molecular subtypes of lung cancer. Nat Rev Clin Oncol 20, 229–249 (2023). https://doi.org/10.1038/s41571-023-00733-6
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41571-023-00733-6
This article is cited by
-
Systematic proteome-wide Mendelian randomization using the human plasma proteome to identify therapeutic targets for lung adenocarcinoma
Journal of Translational Medicine (2024)
-
LTK mutations responsible for resistance to lorlatinib in non-small cell lung cancer harboring CLIP1-LTK fusion
Communications Biology (2024)
-
Lung cancer in patients who have never smoked — an emerging disease
Nature Reviews Clinical Oncology (2024)
-
Current challenges and practical aspects of molecular pathology for non-small cell lung cancers
Virchows Archiv (2024)
-
RIT1 regulates mitosis and promotes proliferation by interacting with SMC3 and PDS5 in hepatocellular carcinoma
Journal of Experimental & Clinical Cancer Research (2023)