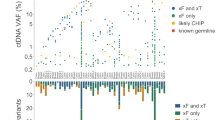
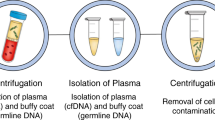
Abstract
Historically, studies of disseminated tumour cells in bone marrow and circulating tumour cells in peripheral blood have provided crucial insights into cancer biology and the metastatic process. More recently, advances in the detection and characterization of circulating tumour DNA (ctDNA) have finally enabled the introduction of liquid biopsy assays into clinical practice. The FDA has already approved several single-gene assays and, more recently, multigene assays to detect genetic alterations in plasma cell-free DNA (cfDNA) for use as companion diagnostics matched to specific molecularly targeted therapies for cancer. These approvals mark a tipping point for the widespread use of liquid biopsy in the clinic, and mostly in patients with advanced-stage cancer. The next frontier for the clinical application of liquid biopsy is likely to be the systemic treatment of patients with ‘ctDNA relapse’, a term we introduce for ctDNA detection prior to imaging-detected relapse after curative-intent therapy for early stage disease. Cancer screening and diagnosis are other potential future applications. In this Perspective, we discuss key issues and gaps in technology, clinical trial methodologies and logistics for the eventual integration of liquid biopsy into the clinical workflow.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
McGranahan, N. & Swanton, C. Clonal heterogeneity and tumor evolution: past, present, and the future. Cell 168, 613–628 (2017).
López, S. et al. Interplay between whole-genome doubling and the accumulation of deleterious alterations in cancer evolution. Nat. Genet. 52, 283–293 (2020).
Yates, L. R. & Campbell, P. J. Evolution of the cancer genome. Nat. Rev. Genet. 13, 795–806 (2012).
Russo, M. et al. Adaptive mutability of colorectal cancers in response to targeted therapies. Science 366, 1473–1480 (2019).
Heitzer, E., Haque, I. S., Roberts, C. E. S. & Speicher, M. R. Current and future perspectives of liquid biopsies in genomics-driven oncology. Nat. Rev. Genet. 20, 71–88 (2019).
Hudson, T. J. et al. International network of cancer genome projects. Nature 464, 993–998 (2010).
Hoadley, K. A. et al. Cell-of-origin patterns dominate the molecular classification of 10,000 tumors from 33 types of cancer. Cell 173, 291–304.e6 (2018).
ICGC/TCGA Pan-Cancer Analysis of Whole Genomes Consortium. Pan-cancer analysis of whole genomes. Nature 578, 82–93 (2020).
Rozenblatt-Rosen, O. et al. The Human Tumor Atlas Network: charting tumor transitions across space and time at single-cell resolution. Cell 181, 236–249 (2020).
Navin, N. et al. Tumour evolution inferred by single-cell sequencing. Nature 472, 90–94 (2011).
Ramalingam, N. & Jeffrey, S. S. Future of liquid biopsies with growing technological and bioinformatics studies: opportunities and challenges in discovering tumor heterogeneity with single-cell level analysis. Cancer J. 24, 104–108 (2018).
Bertucci, F. et al. Genomic characterization of metastatic breast cancers. Nature 569, 560–564 (2019).
Reiter, J. G. et al. An analysis of genetic heterogeneity in untreated cancers. Nat. Rev. Cancer 19, 639–650 (2019).
Hanahan, D. & Coussens, L. M. Accessories to the crime: functions of cells recruited to the tumor microenvironment. Cancer Cell 21, 309–322 (2012).
Pantel, K. & Alix-Panabières, C. Circulating tumour cells in cancer patients: challenges and perspectives. Trends Mol. Med. 16, 398–406 (2010).
Wan, J. C. M. et al. Liquid biopsies come of age: towards implementation of circulating tumour DNA. Nat. Rev. Cancer 17, 223–238 (2017).
Cherry, S. R. et al. Total-body PET: maximizing sensitivity to create new opportunities for clinical research and patient care. J. Nucl. Med. 59, 3–12 (2018).
Ignatiadis, M., Lee, M. & Jeffrey, S. S. Circulating tumor cells and circulating tumor DNA: challenges and opportunities on the path to clinical utility. Clin. Cancer Res. 21, 4786–4800 (2015).
Siravegna, G., Marsoni, S., Siena, S. & Bardelli, A. Integrating liquid biopsies into the management of cancer. Nat. Rev. Clin. Oncol. 14, 531–548 (2017).
Pantel, K. & Alix-Panabières, C. Liquid biopsy and minimal residual disease — latest advances and implications for cure. Nat. Rev. Clin. Oncol. 16, 409–424 (2019).
Cescon, D. W., Bratman, S. V., Chan, S. M. & Siu, L. L. Circulating tumor DNA and liquid biopsy in oncology. Nat. Cancer 1, 276–290 (2020).
Lee, J. S., Park, S. S., Lee, Y. K., Norton, J. A. & Jeffrey, S. S. Liquid biopsy in pancreatic ductal adenocarcinoma: current status of circulating tumor cells and circulating tumor DNA. Mol. Oncol. 13, 1623–1650 (2019).
Teutsch, S. M. et al. The Evaluation of Genomic Applications in Practice and Prevention (EGAPP) initiative: methods of the EGAPP working group. Genet. Med. 11, 3–14 (2009).
Cheng, D. T. et al. Memorial Sloan Kettering-Integrated Mutation Profiling of Actionable Cancer Targets (MSK-IMPACT): a hybridization capture-based next-generation sequencing clinical assay for solid tumor molecular oncology. J. Mol. Diagn. 17, 251–264 (2015).
Zehir, A. et al. Mutational landscape of metastatic cancer revealed from prospective clinical sequencing of 10,000 patients. Nat. Med. 23, 703–713 (2017).
Samstein, R. M. et al. Tumor mutational load predicts survival after immunotherapy across multiple cancer types. Nat. Genet. 51, 202–206 (2019).
US Food and Drug Administration. MSK-IMPACT (Integrated Mutation Profiling of Actionable Cancer Targets). FDA https://www.accessdata.fda.gov/cdrh_docs/pdf17/DEN170058.pdf (2017).
Gao, J. et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci. Signal. 6, pl1 (2013).
Cerami, E. et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2, 401–404 (2012).
Chakravarty, D. et al. OncoKB: a precision oncology knowledge base. JCO Precis. Oncol. 2017, PO.17.00011 (2017).
New York State Department of Health, Wadsworth Center. Memorial Hosp For Cancer and Allied Diseases Dept of Pathology. New York State https://www.wadsworth.org/memorial-hosp-for-cancer-and-allied-diseases-dept-of-pathology-115 (2020).
Razavi, P. et al. High-intensity sequencing reveals the sources of plasma circulating cell-free DNA variants. Nat. Med. 25, 1928–1937 (2019).
Mateo, J. et al. A framework to rank genomic alterations as targets for cancer precision medicine: the ESMO Scale for Clinical Actionability of Molecular Targets (ESCAT). Ann. Oncol. 29, 1895–1902 (2018).
Katsoulakis, E., Duffy, J. E., Hintze, B., Spector, N. L. & Kelley, M. J. Comparison of annotation services for next-generation sequencing in a large-scale precision oncology program. JCO Precis. Oncol. 4, 212–221 (2020).
Wagner, A. H. et al. A harmonized meta-knowledgebase of clinical interpretations of somatic genomic variants in cancer. Nat. Genet. 52, 448–457 (2020).
Van Norman, G. A. Drugs and devices: comparison of European and US approval processes. JACC Basic. Transl. Sci. 1, 399–412 (2016).
Pittella-Silva, F. et al. Plasma or serum: which is preferable for mutation detection in liquid biopsy? Clin. Chem. 66, 946–957 (2020).
Parpart-Li, S. et al. The effect of preservative and temperature on the analysis of circulating tumor DNA. Clin. Cancer Res. 23, 2471–2477 (2017).
Gerber, T. et al. Assessment of pre-analytical sample handling conditions for comprehensive liquid biopsy analysis. J. Mol. Diagn. 22, 1070–1086 (2020).
Salvianti, F. et al. The pre-analytical phase of the liquid biopsy. New Biotechnol. 55, 19–29 (2020).
Grölz, D. et al. Liquid biopsy preservation solutions for standardized pre-analytical workflows — venous whole blood and plasma. Curr. Pathobiol. Rep. 6, 275–286 (2018).
Van Paemel, R. et al. Genome-wide study of the effect of blood collection tubes on the cell-free DNA methylome. Epigenetics https://doi.org/10.1080/15592294.2020.1827714 (2020).
Greytak, S. R. et al. Harmonizing cell-free DNA collection and processing practices through evidence-based guidance. Clin. Cancer Res. 26, 3104–3109 (2020).
Merker, J. D. et al. Circulating tumor DNA analysis in patients with cancer: American Society of Clinical Oncology and College of American Pathologists joint review. J. Clin. Oncol. 36, 1631–1641 (2018).
Andree, K. C., van Dalum, G. & Terstappen, L. W. M. M. Challenges in circulating tumor cell detection by the CellSearch system. Mol. Oncol. 10, 395–407 (2016).
Wong, K. H. K. et al. Whole blood stabilization for the microfluidic isolation and molecular characterization of circulating tumor cells. Nat. Commun. 8, 1733 (2017).
Hodgkinson, C. L. et al. Tumorigenicity and genetic profiling of circulating tumor cells in small-cell lung cancer. Nat. Med. 20, 897–903 (2014).
Yu, M. et al. Ex vivo culture of circulating breast tumor cells for individualized testing of drug susceptibility. Science 345, 216–220 (2014).
Fehm, T. N. et al. Diagnostic leukapheresis for CTC analysis in breast cancer patients: CTC frequency, clinical experiences and recommendations for standardized reporting. Cytometry A 93, 1213–1219 (2018).
Keller, L. & Pantel, K. Unravelling tumour heterogeneity by single-cell profiling of circulating tumour cells. Nat. Rev. Cancer 19, 553–567 (2019).
Cook, L. et al. Does size matter? Comparison of extraction yields for different-sized DNA fragments by seven different routine and four new circulating cell-free extraction methods. J. Clin. Microbiol. 56, e01061-18 (2018).
Meddeb, R., Pisareva, E. & Thierry, A. Guidelines for the preanalytical conditions for analyzing circulating cell-free DNA. Clin. Chem. 65, 623–633 (2019).
Lampignano, R. et al. Multicenter evaluation of circulating cell-free DNA extraction and downstream analyses for the development of standardized (pre)analytical work flows. Clin. Chem. 66, 149–160 (2020).
Febbo, P. G. et al. Minimum technical data elements for liquid biopsy data submitted to public databases. Clin. Pharmacol. Ther. 107, 730–734 (2020).
Foundation for the National Institutes of Health. Biomarkers Consortium – Identification and validation of ctDNA quality control materials. FNIH https://fnih.org/ctdna (2020).
Hao, Y. X. et al. Effectiveness of circulating tumor DNA for detection of KRAS gene mutations in colorectal cancer patients: a meta-analysis. Onco. Targets Ther. 10, 945–953 (2017).
Oxnard, G. R. et al. Noninvasive detection of response and resistance in EGFR-mutant lung cancer using quantitative next-generation genotyping of cell-free plasma DNA. Clin. Cancer Res. 20, 1698–1705 (2014).
Bidard, F.-C. et al. Circulating tumor cells and circulating tumor DNA detection in potentially resectable metastatic colorectal cancer: a prospective ancillary study to the Unicancer PRODIGE-14 trial. Cells 8, 516 (2019).
US Food and Drug Administration. cobas® EGFR Mutation Test v2. FDA https://www.accessdata.fda.gov/cdrh_docs/pdf15/P150047A.pdf (2016).
André, F. et al. Alpelisib for PIK3CA-mutated, hormone receptor-positive advanced breast cancer. N. Engl. J. Med. 380, 1929–1940 (2019).
US Food and Drug Administration. therascreen PIK3CA RGQ PCR kit. FDA https://www.accessdata.fda.gov/cdrh_docs/pdf19/P190004A.pdf (2019).
Kumar, S. et al. Tracking plasma DNA mutation dynamics in estrogen receptor positive metastatic breast cancer with dPCR-SEQ. NPJ Breast Cancer 4, 39 (2018).
Sabari, J. K. et al. A prospective study of circulating tumor DNA to guide matched targeted therapy in lung cancers. J. Natl Cancer Inst. 111, 575–583 (2019).
Clark, T. A. et al. Analytical validation of a hybrid capture-based next-generation sequencing clinical assay for genomic profiling of cell-free circulating tumor DNA. J. Mol. Diagn. 20, 686–702 (2018).
Zill, O. A. et al. The landscape of actionable genomic alterations in cell-free circulating tumor DNA from 21,807 advanced cancer patients. Clin. Cancer Res. 24, 3528–3538 (2018).
Aggarwal, C. et al. Clinical implications of plasma-based genotyping with the delivery of personalized therapy in metastatic non-small cell lung cancer. JAMA Oncol. 5, 173–180 (2019).
Mack, P. C. et al. Spectrum of driver mutations and clinical impact of circulating tumor DNA analysis in non-small cell lung cancer: analysis of over 8000 cases. Cancer 126, 3219–3228 (2020).
Pritchett, M. A. et al. Prospective clinical validation of the InVisionFirst-Lung circulating tumor DNA assay for molecular profiling of patients with advanced nonsquamous non-small-cell lung cancer. JCO Precis. Oncol. 3, PO.18.00299 (2019).
Finzel, A., Sadik, H., Ghitti, G. & Laes, J.-F. The combined analysis of solid and liquid biopsies provides additional clinical information to improve patient care. J. Cancer Metastasis Treat. 4, 21 (2018).
Owonikoko, T. K. et al. Randomized phase II study of paclitaxel plus alisertib versus paclitaxel plus placebo as second-line therapy for SCLC: primary and correlative biomarker analyses. J. Thorac. Oncol. 15, 274–287 (2020).
Leighl, N. B. et al. Clinical utility of comprehensive cell-free DNA analysis to identify genomic biomarkers in patients with newly diagnosed metastatic non-small cell lung cancer. Clin. Cancer Res. 25, 4691–4700 (2019).
Nakamura, Y. et al. Clinical utility of circulating tumor DNA sequencing in advanced gastrointestinal cancer: SCRUM-Japan GI-SCREEN and GOZILA studies. Nat. Med. 26, 1859–1864 (2020).
Razavi, P. et al. Alterations in PTEN and ESR1 promote clinical resistance to alpelisib plus aromatase inhibitors. Nat. Cancer 1, 382–393 (2020).
Kilgour, E., Rothwell, D. G., Brady, G. & Dive, C. Liquid biopsy-based biomarkers of treatment response and resistance. Cancer Cell 37, 485–495 (2020).
Drilon, A. et al. Antitumor activity of crizotinib in lung cancers harboring a MET exon 14 alteration. Nat. Med. 26, 47–51 (2020).
Rothé, F. et al. Plasma circulating tumor DNA as an alternative to metastatic biopsies for mutational analysis in breast cancer. Ann. Oncol. 25, 1959–1965 (2014).
Bachet, J. B. et al. RAS mutation analysis in circulating tumor DNA from patients with metastatic colorectal cancer: the AGEO RASANC prospective multicenter study. Ann. Oncol. 29, 1211–1219 (2018).
Torga, G. & Pienta, K. J. Patient-paired sample congruence between 2 commercial liquid biopsy tests. JAMA Oncol. 4, 868–870 (2018).
Stetson, D. et al. Orthogonal comparison of four plasma NGS tests with tumor suggests technical factors are a major source of assay discordance. JCO Precis. Oncol. 3, 1–9 (2019).
US Food and Drug Administration. Guardant360® CDx. FDA https://www.accessdata.fda.gov/cdrh_docs/pdf20/P200010A.pdf (2020).
Scheerens, H. et al. Current status of companion and complementary diagnostics: strategic considerations for development and launch. Clin. Transl. Sci. 10, 84–92 (2017).
US Food and Drug Administration. FoundationOne® Liquid CDx (F1 Liquid CDx). FDA https://www.accessdata.fda.gov/cdrh_docs/pdf20/P200016A.pdf (2020).
André, F. et al. Comparative genomic hybridisation array and DNA sequencing to direct treatment of metastatic breast cancer: a multicentre, prospective trial (SAFIR01/UNICANCER). Lancet Oncol. 15, 267–274 (2014).
Bettegowda, C. et al. Detection of circulating tumor DNA in early- and late-stage human malignancies. Sci. Transl. Med. 6, 224ra24 (2014).
Abbosh, C. et al. Phylogenetic ctDNA analysis depicts early-stage lung cancer evolution. Nature 545, 446–451 (2017).
Shlush, L. I. Age-related clonal hematopoiesis. Blood 131, 496–504 (2018).
Steensma, D. P. et al. Clonal hematopoiesis of indeterminate potential and its distinction from myelodysplastic syndromes. Blood 126, 9–16 (2015).
Steensma, D. P. & Ebert, B. L. Clonal hematopoiesis as a model for premalignant changes during aging. Exp. Hematol. 83, 48–56 (2020).
Acuna-Hidalgo, R. et al. Ultra-sensitive sequencing identifies high prevalence of clonal hematopoiesis-associated mutations throughout adult life. Am. J. Hum. Genet. 101, 50–64 (2017).
Hu, Y. et al. False-positive plasma genotyping due to clonal hematopoiesis. Clin. Cancer Res. 24, 4437–4443 (2018).
Chan, H. T., Chin, Y. M., Nakamura, Y. & Low, S. K. Clonal hematopoiesis in liquid biopsy: from biological noise to valuable clinical implications. Cancers 12, 2277 (2020).
Jensen, K. et al. Association of clonal hematopoiesis in DNA repair genes with prostate cancer plasma cell-free DNA testing interference. JAMA Oncol. https://doi.org/10.1001/jamaoncol.2020.5161 (2020).
Rothwell, D. G. et al. Utility of ctDNA to support patient selection for early phase clinical trials: the TARGET study. Nat. Med. 25, 738–743 (2019).
Juric, D. et al. Alpelisib + fulvestrant for advanced breast cancer: subgroup analyses from the phase III SOLAR-1 trial [abstract]. Cancer Res 79 (Suppl. 4), GS3-08 (2019).
Turner, N. C. et al. Circulating tumour DNA analysis to direct therapy in advanced breast cancer (plasmaMATCH): a multicentre, multicohort, phase 2a, platform trial. Lancet Oncol. 21, 1296–1308 (2020).
Li, S. et al. Coexistence of EGFR with KRAS, or BRAF, or PIK3CA somatic mutations in lung cancer: a comprehensive mutation profiling from 5125 Chinese cohorts. Br. J. Cancer. 110, 2812–2820 (2014).
De Roock, W., De Vriendt, V., Normanno, N., Ciardiello, F. & Tejpar, S. KRAS, BRAF, PIK3CA, and PTEN mutations: implications for targeted therapies in metastatic colorectal cancer. Lancet Oncol. 12, 594–603 (2011).
Misale, S. et al. Emergence of KRAS mutations and acquired resistance to anti-EGFR therapy in colorectal cancer. Nature 486, 532–536 (2012).
Diaz, L. A. et al. The molecular evolution of acquired resistance to targeted EGFR blockade in colorectal cancers. Nature 486, 537–540 (2012).
Mok, T. S. et al. Osimertinib or platinum-pemetrexed in EGFR T790M-positive lung cancer. N. Engl. J. Med. 376, 629–640 (2017).
Li, B. T. et al. Ultra-deep next-generation sequencing of plasma cell-free DNA in patients with advanced lung cancers: results from the actionable genome consortium. Ann. Oncol. 30, 597–603 (2019).
Heitzer, E. et al. Tumor-associated copy number changes in the circulation of patients with prostate cancer identified through whole-genome sequencing. Genome Med. 5, 30 (2013).
Bourrier, C. et al. Shallow whole-genome sequencing from plasma identifies FGFR1 amplified breast cancers and predicts overall survival. Cancers 12, 1481 (2020).
Murtaza, M. et al. Non-invasive analysis of acquired resistance to cancer therapy by sequencing of plasma DNA. Nature 497, 108–112 (2013).
O’Leary, B. et al. The genetic landscape and clonal evolution of breast cancer resistance to palbociclib plus fulvestrant in the PALOMA-3 trial. Cancer Discov. 8, 1390–1403 (2018).
Antonarakis, E. S. et al. AR-V7 and resistance to enzalutamide and abiraterone in prostate cancer. N. Engl. J. Med. 371, 1028–1038 (2014).
Scher, H. I. et al. Assessment of the validity of nuclear-localized androgen receptor splice variant 7 in circulating tumor cells as a predictive biomarker for castration-resistant prostate cancer. JAMA Oncol. 4, 1179–1186 (2018).
Armstrong, A. J. et al. Prospective multicenter validation of androgen receptor splice variant 7 and hormone therapy resistance in high-risk castration-resistant prostate cancer: the PROPHECY study. in. J. Clin. Oncol. 37, 1120–1129 (2019).
Parikh, A. R. et al. Liquid versus tissue biopsy for detecting acquired resistance and tumor heterogeneity in gastrointestinal cancers. Nat. Med. 25, 1415–1421 (2019).
Pairawan, S. et al. Cell-free circulating tumor DNA variant allele frequency associates with survival in metastatic cancer. Clin. Cancer Res. 26, 1924–1931 (2020).
Beau-Faller, M. et al. Independent prognostic value of ultra-sensitive quantification of tumor pre-treatment T790M subclones in EGFR mutated non-small cell lung cancer (NSCLC) treated by first/second generation TKI, depends on variant allele frequency (VAF): results of the French Cooperative Thoracic Intergroup (IFCT) Biomarkers France project. Lung Cancer 140, 19–26 (2020).
Ma, N. & Jeffrey, S. S. Deciphering cancer clues from blood. Science 367, 1424–1425 (2020).
Ignatiadis, M. et al. Prognostic value of the molecular detection of circulating tumor cells using a multimarker reverse transcription-PCR assay for cytokeratin 19, mammaglobin A, and HER2 in early breast cancer. Clin. Cancer Res. 14, 2593–2600 (2008).
Powell, A. A. et al. Single cell profiling of circulating tumor cells: transcriptional heterogeneity and diversity from breast cancer cell lines. PLoS ONE 7, e33788 (2012).
Lim, S. et al. Liquid biopsy: one cell at a time. NPJ Precis. Oncol. 3, 23 (2019).
Ebright, R. Y. et al. Deregulation of ribosomal protein expression and translation promotes breast cancer metastasis. Science 367, 1468–1473 (2020).
Cabel, L. et al. Clinical potential of circulating tumour DNA in patients receiving anticancer immunotherapy. Nat. Rev. Clin. Oncol. 15, 639–650 (2018).
Hofman, P., Heeke, S., Alix-Panabières, C. & Pantel, K. Liquid biopsy in the era of immuno-oncology: is it ready for prime-time use for cancer patients? Ann. Oncol. 30, 1448–1459 (2019).
Heller, G. et al. Circulating tumor cell number as a response measure of prolonged survival for metastatic castration-resistant prostate cancer: a comparison with prostate-specific antigen across five randomized phase III clinical trials. J. Clin. Oncol. 36, 572–580 (2018).
Cristofanilli, M. et al. Circulating tumor cells, disease progression, and survival in metastatic breast cancer. N. Engl. J. Med. 351, 781–791 (2004).
Bidard, F.-C. et al. Clinical validity of circulating tumour cells in patients with metastatic breast cancer: a pooled analysis of individual patient data. Lancet Oncol. 15, 406–414 (2014).
Moreno, J. G. et al. Circulating tumor cells predict survival in patients with metastatic prostate cancer. Urology 65, 713–718 (2005).
Cohen, S. J. et al. Prognostic significance of circulating tumor cells in patients with metastatic colorectal cancer. Ann. Oncol. 20, 1223–1229 (2009).
Smerage, J. B. et al. Circulating tumor cells and response to chemotherapy in metastatic breast cancer: SWOG S0500. J. Clin. Oncol. 32, 3483–3489 (2014).
Markou, A. et al. PIK3CA mutational status in circulating tumor cells can change during disease recurrence or progression in patients with breast cancer. Clin. Cancer Res. 20, 5823–5834 (2014).
Gasch, C. et al. Frequent detection of PIK3CA mutations in single circulating tumor cells of patients suffering from HER2-negative metastatic breast cancer. Mol. Oncol. 10, 1330–1343 (2016).
Dawson, S. J. et al. Analysis of circulating tumor DNA to monitor metastatic breast cancer. N. Engl. J. Med. 368, 1199–1209 (2013).
Ignatiadis, M. et al. Different prognostic value of cytokeratin-19 mRNA-positive circulating tumor cells according to estrogen receptor and HER2 status in early-stage breast cancer. J. Clin. Oncol. 25, 5194–5202 (2007).
Riethdorf, S. et al. Prognostic impact of circulating tumor cells for breast cancer patients treated in the neoadjuvant “Geparquattro” trial. Clin. Cancer Res. 23, 5384–5393 (2017).
Bidard, F.-C. et al. Circulating tumor cells in breast cancer patients treated by neoadjuvant chemotherapy: a meta-analysis. J. Natl Cancer Inst. 110, 560–567 (2018).
Sparano, J. et al. Association of circulating tumor cells with late recurrence of estrogen receptor-positive breast cancer: a secondary analysis of a randomized clinical trial. JAMA Oncol. 4, 1700–1706 (2018).
Thery, L. et al. Circulating tumor cells in early breast cancer. JNCI Cancer Spectr. 3, pkz026 (2019).
Trapp, E. et al. Presence of circulating tumor cells in high-risk early breast cancer during follow-up and prognosis. J. Natl Cancer Inst. 111, 380–387 (2019).
Diehl, F. et al. Circulating mutant DNA to assess tumor dynamics. Nat. Med. 14, 985–990 (2008).
Garcia-Murillas, I. et al. Mutation tracking in circulating tumor DNA predicts relapse in early breast cancer. Sci. Transl. Med. 7, 302ra133 (2015).
Garcia-Murillas, I. et al. Assessment of molecular relapse detection in early-stage breast cancer. JAMA Oncol. 5, 1473–1478 (2019).
Lawrence, M. S. et al. Mutational heterogeneity in cancer and search for new cancer genes. Nature 499, 214–218 (2013).
Berger, M. F. & Mardis, E. R. The emerging clinical relevance of genomics in cancer medicine. Nat. Rev. Clin. Oncol. 15, 353–365 (2018).
Newman, A. M. et al. Integrated digital error suppression for improved detection of circulating tumor DNA. Nat. Biotechnol. 34, 547–555 (2016).
Wang, T. T. et al. High efficiency error suppression for accurate detection of low-frequency variants. Nucleic Acids Res. 47, e87 (2019).
Zviran, A. et al. Genome-wide cell-free DNA mutational integration enables ultra-sensitive cancer monitoring. Nat. Med. 26, 1114–1124 (2020).
Haque, I. S. & Elemento, O. Challenges in using ctDNA to achieve early detection of cancer. Preprint at bioRxiv https://doi.org/10.1101/237578 (2017).
McDonald, B. R. et al. Personalized circulating tumor DNA analysis to detect residual disease after neoadjuvant therapy in breast cancer. Sci. Transl. Med. 11, eaax7392 (2019).
Coombes, R. C. et al. Personalized detection of circulating tumor DNA antedates breast cancer metastatic recurrence. Clin. Cancer Res. 25, 4255–4263 (2019).
Slamon, D. J. et al. Use of chemotherapy plus a monoclonal antibody against HER2 for metastatic breast cancer that overexpresses HER2. N. Engl. J. Med. 344, 783–792 (2001).
Piccart-Gebhart, M. J. et al. Trastuzumab after adjuvant chemotherapy in HER2-positive breast cancer. N. Engl. J. Med. 353, 1659–1672 (2005).
Romond, E. H. et al. Trastuzumab plus adjuvant chemotherapy for operable HER2-positive breast cancer. N. Engl. J. Med. 353, 1673–1684 (2005).
Gianni, L. et al. Neoadjuvant chemotherapy with trastuzumab followed by adjuvant trastuzumab versus neoadjuvant chemotherapy alone, in patients with HER2-positive locally advanced breast cancer (the NOAH trial): a randomised controlled superiority trial with a parallel HER-negative cohort. Lancet 375, 377–384 (2010).
Von Minckwitz, G. et al. Trastuzumab emtansine for residual invasive HER2-positive breast cancer. N. Engl. J. Med. 380, 617–628 (2019).
Cortazar, P. et al. Pathological complete response and long-term clinical benefit in breast cancer: the CTNeoBC pooled analysis. Lancet 384, 164–172 (2014).
Pan, H. et al. 20-year risks of breast-cancer recurrence after stopping endocrine therapy at 5 years. N. Engl. J. Med. 377, 1836–1846 (2017).
US National Library of Medicine. ClinicalTrials.gov https://clinicaltrials.gov/ct2/show/NCT04567420 (2020).
US National Library of Medicine. ClinicalTrials.gov https://clinicaltrials.gov/ct2/show/NCT04089631 (2020).
US National Library of Medicine. ClinicalTrials.gov https://clinicaltrials.gov/ct2/show/NCT03826758 (2020).
Pantel, K. & Hayes, D. F. Disseminated breast tumour cells: biological and clinical meaning. Nat. Rev. Clin. Oncol. 15, 129–131 (2018).
Naume, B. et al. Clinical outcome with correlation to disseminated tumor cell (DTC) status after DTC-guided secondary adjuvant treatment with docetaxel in early breast cancer. J. Clin. Oncol. 32, 3848–3857 (2014).
Ignatiadis, M. et al. Trastuzumab versus observation for HER2 nonamplified early breast cancer with circulating tumor cells (EORTC 90091-10093, BIG 1-12, Treat CTC): a randomized phase II trial. Ann. Oncol. 29, 1777–1783 (2018).
Smith, M. R. et al. Apalutamide treatment and metastasis-free survival in prostate cancer. N. Engl. J. Med. 378, 1408–1418 (2018).
Hussain, M. et al. Enzalutamide in men with nonmetastatic, castration-resistant prostate cancer. N. Engl. J. Med. 378, 2465–2474 (2018).
Fizazi, K. et al. Darolutamide in nonmetastatic, castration-resistant prostate cancer. N. Engl. J. Med. 380, 1235–1246 (2019).
Gökbuget, N. et al. Blinatumomab for minimal residual disease in adults with B-cell precursor acute lymphoblastic leukemia. Blood 131, 1522–1531 (2018).
Phallen, J. et al. Direct detection of early-stage cancers using circulating tumor DNA. Sci. Transl. Med. 9, eaan2415 (2017).
Bick, A. G. et al. Inherited causes of clonal haematopoiesis in 97,691 whole genomes. Nature 586, 763–768 (2020).
Cohen, J. D. et al. Detection and localization of surgically resectable cancers with a multi-analyte blood test. Science 359, 926–930 (2018).
Shen, S. Y. et al. Sensitive tumour detection and classification using plasma cell-free DNA methylomes. Nature 563, 579–583 (2018).
Hao, X. et al. DNA methylation markers for diagnosis and prognosis of common cancers. Proc. Natl Acad. Sci. USA 114, 7414–7419 (2017).
Cristiano, S. et al. Genome-wide cell-free DNA fragmentation in patients with cancer. Nature 570, 385–389 (2019).
Mouliere, F. et al. Enhanced detection of circulating tumor DNA by fragment size analysis. Sci. Transl. Med. 10, eaat4921 (2018).
Ulz, P. et al. Inference of transcription factor binding from cell-free DNA enables tumor subtype prediction and early detection. Nat. Commun. 10, 4666 (2019).
Liu, M. C. et al. Sensitive and specific multi-cancer detection and localization using methylation signatures in cell-free DNA. Ann. Oncol. 31, 745–759 (2020).
Lennon, A. M. et al. Feasibility of blood testing combined with PET-CT to screen for cancer and guide intervention. Science 369, eabb9601 (2020).
Keller, L., Belloum, Y., Wikman, H. & Pantel, K. Clinical relevance of blood-based ctDNA analysis: mutation detection and beyond. Br. J. Cancer https://doi.org/10.1038/s41416-020-01047-5 (2020).
US Preventive Services Task Force. Prostate cancer screening. USPSTF https://www.uspreventiveservicestaskforce.org/uspstf/recommendation/prostate-cancer-screening (2018).
Paik, S. et al. A multigene assay to predict recurrence of tamoxifen-treated, node-negative breast cancer. N. Engl. J. Med. 351, 2817–2826 (2004).
Frampton, G. M. et al. Development and validation of a clinical cancer genomic profiling test based on massively parallel DNA sequencing. Nat. Biotechnol. 31, 1023–1031 (2013).
Van De Haar, J., Hoes, L. & Voest, E. Advancing molecular tumour boards: highly needed to maximise the impact of precision medicine. ESMO Open 4, e000516 (2019).
Topol, E. J. High-performance medicine: the convergence of human and artificial intelligence. Nat. Med. 25, 44–56 (2019).
Zeune, L. L. et al. How to agree on a CTC: evaluating the consensus in circulating tumor cell scoring. Cytometry A 93, 1202–1206 (2018).
Iyer, A. et al. Integrative analysis and machine learning based characterization of single circulating tumor cells. J. Clin. Med. 9, 1206 (2020).
Ota, S. et al. Ghost cytometry. Science 360, 1246–1251 (2018).
Wan, N. et al. Machine learning enables detection of early-stage colorectal cancer by whole-genome sequencing of plasma cell-free DNA. BMC Cancer 19, 832 (2019).
Camacho, D. M., Collins, K. M., Powers, R. K., Costello, J. C. & Collins, J. J. Next-generation machine learning for biological networks. Cell 173, 1581–1592 (2018).
Chabon, J. J. et al. Integrating genomic features for non-invasive early lung cancer detection. Nature 580, 245–251 (2020).
Menetski, J. P. et al. The Foundation for the National Institutes of Health Biomarkers Consortium: past accomplishments and new strategic direction. Clin. Pharmacol. Ther. 105, 829–843 (2019).
Xu, H. et al. A comparison of EGFR mutation status in tissue and plasma cell-free DNA detected by ADx-ARMS in advanced lung adenocarcinoma patients. Transl. Lung Cancer Res. 8, 135–143 (2019).
Budd, G. T. et al. Circulating tumor cells versus imaging–predicting overall survival in metastatic breast cancer. Clin. Cancer Res. 12, 6403–6409 (2006).
Eisenhauer, E. A. et al. New Response Evaluation Criteria in Solid Tumours: revised RECIST guideline (version 1.1). Eur. J. Cancer 45, 228–247 (2009).
Nelson, H. D. et al. Effectiveness of breast cancer screening: systematic review and meta-analysis to update the 2009 US Preventive Services Task Force recommendation. Ann. Intern. Med. 164, 244–255 (2016).
Ilic, D. et al. Prostate cancer screening with prostate-specific antigen (PSA) test: a systematic review and meta-analysis. BMJ 362, k3519 (2018).
Olsson, E. et al. Serial monitoring of circulating tumor DNA in patients with primary breast cancer for detection of occult metastatic disease. EMBO Mol. Med. 7, 1034–1047 (2015).
Riva, F. et al. Patient-specific circulating tumor DNA detection during neoadjuvant chemotherapy in triple-negative breast cancer. Clin. Chem. 63, 691–699 (2017).
Chen, Y. H. et al. Next-generation sequencing of circulating tumor DNA to predict recurrence in triple-negative breast cancer patients with residual disease after neoadjuvant chemotherapy. NPJ Breast Cancer 3, 24 (2017).
Rothé, F. et al. Circulating tumor DNA in HER2-amplified breast cancer: a translational research substudy of the NeoALTTO phase III trial. Clin. Cancer Res. 25, 3581–3588 (2019).
Zhang, X. et al. Parallel analyses of somatic mutations in plasma circulating tumor DNA (ctDNA) and matched tumor tissues in early-stage breast cancer. Clin. Cancer Res. 25, 6546–6553 (2019).
Radovich, M. et al. Association of circulating tumor DNA and circulating tumor cells after neoadjuvant chemotherapy with disease recurrence in patients with triple-negative breast cancer: preplanned secondary analysis of the BRE12-158 randomized clinical trial. JAMA Oncol. 6, 1410–1415 (2020).
Scher, H. I. et al. Association of AR-V7 on circulating tumor cells as a treatment-specific biomarker with outcomes and survival in castration-resistant prostate cancer. JAMA Oncol. 2, 1441–1449 (2016).
Acknowledgements
The work of M.I. is supported by Les Amis de Bordet and Fondation Contre le Cancer. The work of G.W.S. is supported in part by the Susan G. Komen Foundation. The work of S.S.J. is supported in part by the John and Marva Warnock Research Fund, Natalie and Vladimir Ermakoff, and the Stanford Catalyst for Collaborative Solutions, Stanford School of Engineering.
Author information
Authors and Affiliations
Contributions
All authors contributed to all aspects of manuscript preparation.
Corresponding authors
Ethics declarations
Competing interests
M.I. has received consultancy fees from Celgene, Novartis, Pfizer, Seattle Genetics and Tesaro, and travel grants from Amgen and Pfizer. The institution of M.I. has received research grants from Menarini Silicon Biosystems and Natera. G.W.S. is a member of the Board of Directors of Tessa Therapeutics and of the Scientific Advisory Boards of Syndax and Verseau Therapeutics. S.S.J. serves as a scientific advisor for Quantumcyte and Ravel Biotechnology.
Additional information
Peer review information
Nature Reviews Clinical Oncology thanks R. Rosell; F.-C. Bidard, who co-reviewed with L. Cabel; and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Related links
Blood Profiling Atlas in Cancer (BloodPAC): https://www.bloodpac.org/
CANCER-ID: https://www.cancer-id.eu/
cBioPortal: https://www.cbioportal.org/index.do
European Liquid Biopsy Society (ELBS): http://www.elbs.eu
OncoKB: https://www.oncokb.org/
OncoPrint: https://bioc.ism.ac.jp/packages/3.2/bioc/vignettes/ComplexHeatmap/inst/doc/s8.oncoprint.html#toc_0
Rights and permissions
About this article
Cite this article
Ignatiadis, M., Sledge, G.W. & Jeffrey, S.S. Liquid biopsy enters the clinic — implementation issues and future challenges. Nat Rev Clin Oncol 18, 297–312 (2021). https://doi.org/10.1038/s41571-020-00457-x
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41571-020-00457-x
This article is cited by
-
Optical nanomaterial-based detection of biomarkers in liquid biopsy
Journal of Hematology & Oncology (2024)
-
Soluble Periostin is a potential surveillance biomarker for early and long-term response to chemotherapy in advanced breast cancer
Cancer Cell International (2024)
-
Integrating cfDNA liquid biopsy and organoid-based drug screening reveals PI3K signaling as a promising therapeutic target in colorectal cancer
Journal of Translational Medicine (2024)
-
Decoding the glycoproteome: a new frontier for biomarker discovery in cancer
Journal of Hematology & Oncology (2024)
-
Liquid biopsy in T-cell lymphoma: biomarker detection techniques and clinical application
Molecular Cancer (2024)