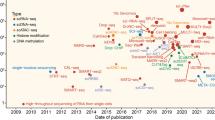
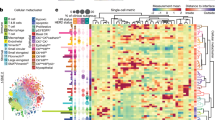
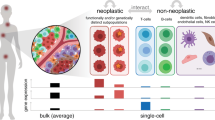
Abstract
Advances in molecular biology, microfluidics and bioinformatics have empowered the study of thousands or even millions of individual cells from malignant tumours at the single-cell level of resolution. This high-dimensional, multi-faceted characterization of the genomic, transcriptomic, epigenomic and proteomic features of the tumour and/or the associated immune and stromal cells enables the dissection of tumour heterogeneity, the complex interactions between tumour cells and their microenvironment, and the details of the evolutionary trajectory of each tumour. Single-cell transcriptomics, the ability to track individual T cell clones through paired sequencing of the T cell receptor genes and high-dimensional single-cell spatial analysis are all areas of particular relevance to immuno-oncology. Multidimensional biomarker signatures will increasingly be crucial to guiding clinical decision-making in each patient with cancer. High-dimensional single-cell technologies are likely to provide the resolution and richness of data required to generate such clinically relevant signatures in immuno-oncology. In this Perspective, we describe advances made using transformative single-cell analysis technologies, especially in relation to clinical response and resistance to immunotherapy, and discuss the growing utility of single-cell approaches for answering important research questions.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
References
Sharma, P., Hu-Lieskovan, S., Wargo, J. A. & Ribas, A. Primary, adaptive, and acquired resistance to cancer immunotherapy. Cell 168, 707–723 (2017).
Iorgulescu, J. B., Braun, D., Oliveira, G., Keskin, D. B. & Wu, C. J. Acquired mechanisms of immune escape in cancer following immunotherapy. Genome Med. 10, 87 (2018).
Wang, L., Livak, K. J. & Wu, C. J. High-dimension single-cell analysis applied to cancer. Mol. Aspects Med. 59, 70–84 (2018).
Finotello, F. & Eduati, F. Multi-omics profiling of the tumor microenvironment: paving the way to precision immuno-oncology. Front. Oncol. 8, 430 (2018).
Gomes, T., Teichmann, S. A. & Talavera-López, C. Immunology driven by large-scale single-cell sequencing. Trends Immunol. 40, 1011–1021 (2019).
Tang, X., Huang, Y., Lei, J., Luo, H. & Zhu, X. The single-cell sequencing: new developments and medical applications. Cell Biosci. 9, 53 (2019).
Chen, G., Ning, B. & Shi, T. Single-cell RNA-Seq technologies and related computational data analysis. Front. Genet. 10, 317 (2019).
Qi, R., Ma, A., Ma, Q. & Zou, Q. Clustering and classification methods for single-cell RNA-sequencing data. Brief. Bioinform. 21, 1196–1208 (2019).
Luecken, M. D. & Theis, F. J. Current best practices in single-cell RNA-seq analysis: a tutorial. Mol. Syst. Biol. 15, e8746 (2019).
Saelens, W., Cannoodt, R., Todorov, H. & Saeys, Y. A comparison of single-cell trajectory inference methods. Nat. Biotechnol. 37, 547–554 (2019).
Lähnemann, D. et al. Eleven grand challenges in single-cell data science. Genome Biol. 21, 31 (2020).
Svensson, V., Vento-Tormo, R. & Teichmann, S. A. Exponential scaling of single-cell RNA-seq in the past decade. Nat. Protoc. 13, 599–604 (2018).
Regev, A. et al. The human cell atlas. eLife 6, e27041 (2017).
Plasschaert, L. W. et al. A single-cell atlas of the airway epithelium reveals the CFTR-rich pulmonary ionocyte. Nature 560, 377–381 (2018).
Villani, A.-C. et al. Single-cell RNA-seq reveals new types of human blood dendritic cells, monocytes, and progenitors. Science 356, eaah4573 (2017).
Filbin, M. G. et al. Developmental and oncogenic programs in H3K27M gliomas dissected by single-cell RNA-seq. Science 360, 331–335 (2018).
Tirosh, I. et al. Dissecting the multicellular ecosystem of metastatic melanoma by single-cell RNA-seq. Science 352, 189–196 (2016).
Jerby-Arnon, L. et al. A cancer cell program promotes T cell exclusion and resistance to checkpoint blockade. Cell 175, 984–997.e24 (2018).
Puram, S. V. et al. Single-cell transcriptomic analysis of primary and metastatic tumor ecosystems in head and neck cancer. Cell 171, 1611–1624.e24 (2017).
Izar, B. et al. A single-cell landscape of high-grade serous ovarian cancer. Nat. Med. 26, 1271–1279 (2020).
Tirosh, I. et al. Single-cell RNA-seq supports a developmental hierarchy in human oligodendroglioma. Nature 539, 309–313 (2016).
Venteicher, A. S. et al. Decoupling genetics, lineages, and microenvironment in IDH-mutant gliomas by single-cell RNA-seq. Science 355, eaai8478 (2017).
Neftel, C. et al. An integrative model of cellular states, plasticity, and genetics for glioblastoma. Cell 178, 835–849.e21 (2019).
Young, M. D. et al. Single-cell transcriptomes from human kidneys reveal the cellular identity of renal tumors. Science 361, 594–599 (2018).
Szabo, P. A. et al. Single-cell transcriptomics of human T cells reveals tissue and activation signatures in health and disease. Nat. Commun. 10, 4706 (2019).
Lambrechts, D. et al. Phenotype molding of stromal cells in the lung tumor microenvironment. Nat. Med. 24, 1277–1289 (2018).
Zhang, L. et al. Lineage tracking reveals dynamic relationships of T cells in colorectal cancer. Nature 564, 268–272 (2018).
Azizi, E. et al. Single-cell map of diverse immune phenotypes in the breast tumor microenvironment. Cell 174, 1293–1308.e36 (2018).
Sade-Feldman, M. et al. Defining T cell states associated with response to checkpoint immunotherapy in melanoma. Cell 175, 998–1013.e20 (2018).
Efremova, M., Vento-Tormo, M., Teichmann, S. A. & Vento-Tormo, R. CellPhoneDB: inferring cell-cell communication from combined expression of multi-subunit ligand-receptor complexes. Nat. Protoc. 15, 1484–1506 (2020).
Cabello-Aguilar, S. et al. SingleCellSignalR: inference of intercellular networks from single-cell transcriptomics. Nucleic Acids Res. 48, e55 (2020).
Zhang, Q. et al. Landscape and dynamics of single immune cells in hepatocellular carcinoma. Cell 179, 829–845.e20 (2019).
Chubb, J. R., Trcek, T., Shenoy, S. M. & Singer, R. H. Transcriptional pulsing of a developmental gene. Curr. Biol. 16, 1018–1025 (2006).
Dar, R. D. et al. Transcriptional burst frequency and burst size are equally modulated across the human genome. Proc. Natl Acad. Sci. USA 109, 17454–17459 (2012).
Deng, Q., Ramsköld, D., Reinius, B. & Sandberg, R. Single-cell RNA-seq reveals dynamic, random monoallelic gene expression in mammalian cells. Science 343, 193–196 (2014).
Reinius, B. & Sandberg, R. Random monoallelic expression of autosomal genes: stochastic transcription and allele-level regulation. Nat. Rev. Genet. 16, 653–664 (2015).
Patel, A. P. et al. Single-cell RNA-seq highlights intratumoral heterogeneity in primary glioblastoma. Science 344, 1396–1401 (2014).
Müller, S. et al. Single-cell sequencing maps gene expression to mutational phylogenies in PDGF- and EGF-driven gliomas. Mol. Syst. Biol. 12, 889 (2016).
Rodriguez-Meira, A. et al. Unravelling intratumoral heterogeneity through high-sensitivity single-cell mutational analysis and parallel RNA sequencing. Mol. Cell 73, 1292–1305.e8 (2019).
Nam, A. S. et al. Somatic mutations and cell identity linked by genotyping of transcriptomes. Nature 571, 355–360 (2019).
van Galen, P. et al. Single-cell RNA-Seq reveals AML hierarchies relevant to disease progression and immunity. Cell 176, 1265–1281.e24 (2019).
van den Brink, S. C. et al. Single-cell sequencing reveals dissociation-induced gene expression in tissue subpopulations. Nat. Methods 14, 935–936 (2017).
Massoni-Badosa, R. et al. Sampling time-dependent artifacts in single-cell genomics studies. Genome Biol. 21, 112 (2020).
Slyper, M. et al. A single-cell and single-nucleus RNA-Seq toolbox for fresh and frozen human tumors. Nat. Med. 26, 792–802 (2020).
Habib, N. et al. Massively parallel single-nucleus RNA-seq with DroNc-seq. Nat. Methods 14, 955–958 (2017).
Bakken, T. E. et al. Single-nucleus and single-cell transcriptomes compared in matched cortical cell types. PLoS ONE 13, e0209648 (2018).
Wu, H., Kirita, Y., Donnelly, E. L. & Humphreys, B. D. Advantages of single-nucleus over single-cell RNA sequencing of adult kidney: rare cell types and novel cell states revealed in fibrosis. J. Am. Soc. Nephrol. 30, 23–32 (2019).
Liang, Q. et al. Single-nuclei RNA-seq on human retinal tissue provides improved transcriptome profiling. Nat. Commun. 10, 5743 (2019).
Tran, H. T. N. et al. A benchmark of batch-effect correction methods for single-cell RNA sequencing data. Genome Biol. 21, 12 (2020).
Zhang, L. & Vertes, A. Single-cell mass spectrometry approaches to explore cellular heterogeneity. Angew. Chem. Int. Ed. Engl. 57, 4466–4477 (2018).
Löfblom, J., Frejd, F. Y. & Ståhl, S. Non-immunoglobulin based protein scaffolds. Curr. Opin. Biotechnol. 22, 843–848 (2011).
Banta, S., Dooley, K. & Shur, O. Replacing antibodies: engineering new binding proteins. Annu. Rev. Biomed. Eng. 15, 93–113 (2013).
Reverdatto, S., Burz, D. S. & Shekhtman, A. Peptide aptamers: development and applications. Curr. Top. Med. Chem. 15, 1082–1101 (2015).
Ståhl, S. et al. Affibody molecules in biotechnological and medical applications. Trends Biotechnol. 35, 691–712 (2017).
Zhou, Z., Liu, M. & Jiang, J. The potential of aptamers for cancer research. Anal. Biochem. 549, 91–95 (2018).
Spitzer, M. H. & Nolan, G. P. Mass cytometry: single cells, many features. Cell 165, 780–791 (2016).
Wagner, J. et al. A single-cell atlas of the tumor and immune ecosystem of human breast cancer. Cell 177, 1330–1345.e18 (2019).
Fisher, J. et al. Engineering γδT cells limits tonic signaling associated with chimeric antigen receptors. Sci. Signal. 12, eaax1872 (2019).
Tsai, A. G. et al. Multiplexed single-cell morphometry for hematopathology diagnostics. Nat. Med. 26, 408–417 (2020).
Krieg, C. et al. High-dimensional single-cell analysis predicts response to anti-PD-1 immunotherapy. Nat. Med. 24, 144–153 (2018).
Gide, T. N. et al. Distinct immune cell populations define response to anti-PD-1 monotherapy and anti-PD-1/anti-CTLA-4 combined therapy. Cancer Cell 35, 238–255.e6 (2019).
Hennig, C., Adams, N. & Hansen, G. A versatile platform for comprehensive chip-based explorative cytometry. Cytometry A 75, 362–370 (2009).
Teo, J. et al. A preliminary study for the assessment of PD-L1 and PD-L2 on circulating tumor cells by microfluidic-based chipcytometry. Future Sci. OA 3, FSO244 (2017).
Shi, Q. et al. Single-cell proteomic chip for profiling intracellular signaling pathways in single tumor cells. Proc. Natl Acad. Sci. USA 109, 419–424 (2012).
Su, Y. et al. Multi-omic single-cell snapshots reveal multiple independent trajectories to drug tolerance in a melanoma cell line. Nat. Commun. 11, 2345 (2020).
Liu, D., Paczkowski, P., Mackay, S., Ng, C. & Zhou, J. Single-cell multiplexed proteomics on the IsoLight resolves cellular functional heterogeneity to reveal clinical responses of cancer patients to immunotherapies. Methods Mol. Biol. 2055, 413–431 (2020).
Parisi, G. et al. Persistence of adoptively transferred T cells with a kinetically engineered IL-2 receptor agonist. Nat. Commun. 11, 660 (2020).
Gawad, C., Koh, W. & Quake, S. R. Single-cell genome sequencing: current state of the science. Nat. Rev. Genet. 17, 175–188 (2016).
Pellegrino, M. et al. High-throughput single-cell DNA sequencing of acute myeloid leukemia tumors with droplet microfluidics. Genome Res. 28, 1345–1352 (2018).
McMahon, C. M. et al. Clonal selection with RAS pathway activation mediates secondary clinical resistance to selective FLT3 inhibition in acute myeloid leukemia. Cancer Discov. 9, 1050–1063 (2019).
Xu, L. et al. Clonal evolution and changes in two AML patients detected with a novel single-cell DNA sequencing platform. Sci. Rep. 9, 11119 (2019).
Ediriwickrema, A. et al. Single-cell mutational profiling enhances the clinical evaluation of AML MRD. Blood Adv. 4, 943–952 (2020).
DiNardo, C. D. et al. Molecular patterns of response and treatment failure after frontline venetoclax combinations in older patients with AML. Blood 135, 791–803 (2020).
Choe, S. et al. Molecular mechanisms mediating relapse following ivosidenib monotherapy in IDH1-mutant relapsed or refractory AML. Blood Adv. 4, 1894–1905 (2020).
Kelsey, G., Stegle, O. & Reik, W. Single-cell epigenomics: recording the past and predicting the future. Science 358, 69–75 (2017).
Shema, E., Bernstein, B. E. & Buenrostro, J. D. Single-cell and single-molecule epigenomics to uncover genome regulation at unprecedented resolution. Nat. Genet. 51, 19–25 (2019).
Ludwig, C. H. & Bintu, L. Mapping chromatin modifications at the single cell level. Development 146, dev170217 (2019).
Satpathy, A. T. et al. Massively parallel single-cell chromatin landscapes of human immune cell development and intratumoral T cell exhaustion. Nat. Biotechnol. 37, 925–936 (2019).
Cao, J. et al. Joint profiling of chromatin accessibility and gene expression in thousands of single cells. Science 361, 1380–1385 (2018).
Reyes, M., Billman, K., Hacohen, N. & Blainey, P. C. Simultaneous profiling of gene expression and chromatin accessibility in single cells. Adv. Biosys. 3, 1900065 (2019).
Gaiti, F. et al. Epigenetic evolution and lineage histories of chronic lymphocytic leukaemia. Nature 569, 576–580 (2019).
Kaya-Okur, H. S. et al. CUT&Tag for efficient epigenomic profiling of small samples and single cells. Nat. Commun. 10, 1930 (2019).
Stuart, T. & Satija, R. Integrative single-cell analysis. Nat. Rev. Genet. 20, 257–272 (2019).
Peng, A., Mao, X., Zhong, J., Fan, S. & Hu, Y. Single-cell multi-omics and its prospective application in cancer biology. Proteomics 20, 1900271 (2020).
Stoeckius, M. et al. Simultaneous epitope and transcriptome measurement in single cells. Nat. Methods 14, 865–868 (2017).
Peterson, V. M. et al. Multiplexed quantification of proteins and transcripts in single cells. Nat. Biotechnol. 35, 936–939 (2017).
Assarsson, E. et al. Homogenous 96-plex PEA immunoassay exhibiting high sensitivity, specificity, and excellent scalability. PLoS ONE 9, e95192 (2014).
Miles, L. A. et al. Single-cell mutation analysis of clonal evolution in myeloid malignancies. Nature https://doi.org/10.1038/s41586-020-2864-x (2020).
Ludwig, L. S. et al. Lineage tracing in humans enabled by mitochondrial mutations and single-cell genomics. Cell 176, 1325–1339.e22 (2019).
Xu, J. et al. Single-cell lineage tracing by endogenous mutations enriched in transposase accessible mitochondrial DNA. eLife 8, e45105 (2019).
Davis, M. M. T cell receptor gene diversity and selection. Annu. Rev. Biochem. 59, 475–496 (1990).
Yang, J. C. & Rosenberg, S. A. Adoptive T-cell therapy for cancer. Adv. Immunol. 130, 279–294 (2016).
Simoni, Y. et al. Bystander CD8+ T cells are abundant and phenotypically distinct in human tumour infiltrates. Nature 557, 575–579 (2018).
Scheper, W. et al. Low and variable tumor reactivity of the intratumoral TCR repertoire in human cancers. Nat. Med. 25, 89–94 (2019).
Wang, G. C., Dash, P., McCullers, J. A., Doherty, P. C. & Thomas, P. G. T cell receptor αβ diversity inversely correlates with pathogen-specific antibody levels in human cytomegalovirus infection. Sci. Transl Med. 4, 128ra42 (2012).
Han, A., Glanville, J., Hansmann, L. & Davis, M. M. Linking T-cell receptor sequence to functional phenotype at the single-cell level. Nat. Biotechnol. 32, 684–692 (2014).
Dash, P., Wang, G. C. & Thomas, P. G. Single-cell analysis of T-cell receptor αβ repertoire. Methods Mol. Biol. 1343, 181–197 (2015).
Li, S. et al. RNase H-dependent PCR-enabled T-cell receptor sequencing for highly specific and efficient targeted sequencing of T-cell receptor mRNA for single-cell and repertoire analysis. Nat. Protoc. 14, 2571–2594 (2019).
Kobayashi, E. et al. A new cloning and expression system yields and validates TCRs from blood lymphocytes of patients with cancer within 10 days. Nat. Med. 19, 1542–1546 (2013).
Guo, X.-Z. J. et al. Rapid cloning, expression, and functional characterization of paired αβ and γδ T-cell receptor chains from single-cell analysis. Mol. Ther. Methods Clin. Dev. 3, 15054 (2016).
Hu, Z. et al. A cloning and expression system to probe T-cell receptor specificity and assess functional avidity to neoantigens. Blood 132, 1911–1921 (2018).
Kula, T. et al. T-Scan: a genome-wide method for the systematic discovery of T cell epitopes. Cell 178, 1016–1028.e13 (2019).
Zhang, S.-Q. et al. High-throughput determination of the antigen specificities of T cell receptors in single cells. Nat. Biotechnol. 36, 1156–1159 (2018).
Segaliny, A. I. et al. Functional TCR T cell screening using single-cell droplet microfluidics. Lab Chip 18, 3733–3749 (2018).
Keskin, D. B. et al. Neoantigen vaccine generates intratumoral T cell responses in phase Ib glioblastoma trial. Nature 565, 234–239 (2019).
Yost, K. E. et al. Clonal replacement of tumor-specific T cells following PD-1 blockade. Nat. Med. 25, 1251–1259 (2019).
Durante, M. A. et al. Single-cell analysis reveals new evolutionary complexity in uveal melanoma. Nat. Commun. 11, 496 (2020).
Tanno, H. et al. A facile technology for the high-throughput sequencing of the paired VH:VL and TCRβ:TCRα repertoires. Sci. Adv. 6, eaay9093 (2020).
Coons, A. H., Creech, H. J. & Jones, R. N. Immunological properties of an antibody containing a fluorescent group. Exp. Biol. Med. 47, 200–202 (1941).
Gall, J. G. & Pardue, M. L. Formation and detection of RNA-DNA hybrid molecules in cytological preparations. Proc. Natl Acad. Sci. USA 63, 378–383 (1969).
Lu, S. et al. Comparison of biomarker modalities for predicting response to PD-1/PD-L1 checkpoint blockade: a systematic review and meta-analysis. JAMA Oncol. 5, 1195–1204 (2019).
Johnson, D. B. et al. Quantitative spatial profiling of PD-1/PD-L1 interaction and HLA-DR/IDO-1 predicts improved outcomes of anti-PD-1 therapies in metastatic melanoma. Clin. Cancer Res. 24, 5250–5260 (2018).
Jackson, H. W. et al. The single-cell pathology landscape of breast cancer. Nature 578, 615–620 (2020).
Bosisio, F. M. et al. Functional heterogeneity of lymphocytic patterns in primary melanoma dissected through single-cell multiplexing. eLife 9, e53008 (2020).
Helmink, B. A. et al. B cells and tertiary lymphoid structures promote immunotherapy response. Nature 577, 549–555 (2020).
Cabrita, R. et al. Tertiary lymphoid structures improve immunotherapy and survival in melanoma. Nature 577, 561–565 (2020).
Xin Yu, J. et al. Trends in clinical development for PD-1/PD-L1 inhibitors. Nat. Rev. Drug Discov. 19, 163–164 (2020).
McGranahan, N. et al. Clonal neoantigens elicit T cell immunoreactivity and sensitivity to immune checkpoint blockade. Science 351, 1463–1469 (2016).
Reuben, A. et al. Genomic and immune heterogeneity are associated with differential responses to therapy in melanoma. NPJ Genom. Med. 2, 10 (2017).
Rozenblatt-Rosen, O. et al. The Human Tumor Atlas Network: charting tumor transitions across space and time at single-cell resolution. Cell 181, 236–249 (2020).
Newman, A. M. et al. Determining cell type abundance and expression from bulk tissues with digital cytometry. Nat. Biotechnol. 37, 773–782 (2019).
Maïno, N. et al. A microfluidic platform towards automated multiplexed in situ sequencing. Sci. Rep. 9, 3542 (2019).
Maus, M. V. et al. Adoptive immunotherapy for cancer or viruses. Annu. Rev. Immunol. 32, 189–225 (2014).
Martin-Liberal, J. et al. The expanding role of immunotherapy. Cancer Treat. Rev. 54, 74–86 (2017).
Sanghera, C. & Sanghera, R. Immunotherapy – strategies for expanding its role in the treatment of all major tumor sites. Cureus 11, e5938 (2019).
Kieffer, Y. et al. Single-cell analysis reveals fibroblast clusters linked to immunotherapy resistance in cancer. Cancer Discov. 10, 1330–1351 (2020).
Dominguez, C. X. et al. Single-cell RNA sequencing reveals stromal evolution into LRRC15+ myofibroblasts as a determinant of patient response to cancer immunotherapy. Cancer Discov. 10, 232–253 (2020).
Guo, X. et al. Global characterization of T cells in non-small-cell lung cancer by single-cell sequencing. Nat. Med. 24, 978–985 (2018).
Zheng, C. et al. Landscape of infiltrating T cells in liver cancer revealed by single-cell sequencing. Cell 169, 1342–1356.e16 (2017).
Zhang, L. et al. Single-cell analyses inform mechanisms of myeloid-targeted therapies in colon cancer. Cell 181, 442–459.e29 (2020).
Witkowski, M. T. et al. Extensive remodeling of the immune microenvironment in B cell acute lymphoblastic leukemia. Cancer Cell 37, 867–882.e12 (2020).
House, I. G. et al. Macrophage-derived CXCL9 and CXCL10 are required for antitumor immune responses following immune checkpoint blockade. Clin. Cancer Res. 26, 487–504 (2020).
Livak, K. J. in Field Guidelines for Genetic Experimental Designs in High-Throughput Sequencing (eds Aransay, A. M. & Lavín Trueba, J. L.) 343–365 (Springer, 2016).
Gerdes, M. J. et al. Highly multiplexed single-cell analysis of formalin-fixed, paraffin-embedded cancer tissue. Proc. Natl Acad. Sci. USA 110, 11982–11987 (2013).
Lin, J.-R. et al. Highly multiplexed immunofluorescence imaging of human tissues and tumors using t-CyCIF and conventional optical microscopes. eLife 7, e31657 (2018).
Bolognesi, M. M. et al. Multiplex staining by sequential immunostaining and antibody removal on routine tissue sections. J. Histochem. Cytochem. 65, 431–444 (2017).
Cattoretti, G., Bosisio, F. M., Marcelis, L. & Bolognesi, M. M. Multiple iterative labeling by antibody neodeposition (MILAN). Protocol Exchange https://doi.org/10.21203/rs.2.1646/v5 (2019).
Eng, J. et al. Cyclic multiplexed-immunofluorescence (cmIF), a highly multiplexed method for single-cell analysis. Methods Mol. Biol. 2055, 521–562 (2020).
Consentius, C. et al. In situ detection of CD73+ CD90+ CD105+ lineage: mesenchymal stromal cells in human placenta and bone marrow specimens by chipcytometry. Cytometry A 93, 889–893 (2018).
Angelo, M. et al. Multiplexed ion beam imaging of human breast tumors. Nat. Med. 20, 436–442 (2014).
Keren, L. et al. MIBI-TOF: A multiplexed imaging platform relates cellular phenotypes and tissue structure. Sci. Adv. 5, eaax5851 (2019).
Giesen, C. et al. Highly multiplexed imaging of tumor tissues with subcellular resolution by mass cytometry. Nat. Methods 11, 417–422 (2014).
Ramaglia, V. et al. Multiplexed imaging of immune cells in staged multiple sclerosis lesions by mass cytometry. eLife 8, e48051 (2019).
Ijsselsteijn, M. E., van der Breggen, R., Farina Sarasqueta, A., Koning, F. & de Miranda, N. F. C. C. A 40-marker panel for high dimensional characterization of cancer immune microenvironments by imaging mass cytometry. Front. Immunol. 10, 2534 (2019).
Goltsev, Y. et al. Deep profiling of mouse splenic architecture with CODEX multiplexed imaging. Cell 174, 968–981.e15 (2018).
Chen, K. H., Boettiger, A. N., Moffitt, J. R., Wang, S. & Zhuang, X. RNA imaging. Spatially resolved, highly multiplexed RNA profiling in single cells. Science 348, aaa6090 (2015).
Eng, C.-H. L. et al. Transcriptome-scale super-resolved imaging in tissues by RNA seqFISH. Nature 568, 235–239 (2019).
Ståhl, P. L. et al. Visualization and analysis of gene expression in tissue sections by spatial transcriptomics. Science 353, 78–82 (2016).
Thrane, K., Eriksson, H., Maaskola, J., Hansson, J. & Lundeberg, J. Spatially resolved transcriptomics enables dissection of genetic heterogeneity in stage III cutaneous malignant melanoma. Cancer Res. 78, 5970–5979 (2018).
Rodriques, S. G. et al. Slide-seq: a scalable technology for measuring genome-wide expression at high spatial resolution. Science 363, 1463–1467 (2019).
Vickovic, S. et al. High-definition spatial transcriptomics for in situ tissue profiling. Nat. Methods 16, 987–990 (2019).
Lee, J. H. et al. Highly multiplexed subcellular RNA sequencing in situ. Science 343, 1360–1363 (2014).
Lee, J. H. et al. Fluorescent in situ sequencing (FISSEQ) of RNA for gene expression profiling in intact cells and tissues. Nat. Protoc. 10, 442–458 (2015).
Wang, X. et al. Three-dimensional intact-tissue sequencing of single-cell transcriptional states. Science 361, eaat5691 (2018).
Decalf, J., Albert, M. L. & Ziai, J. New tools for pathology: a user’s review of a highly multiplexed method for in situ analysis of protein and RNA expression in tissue. J. Pathol. 247, 650–661 (2019).
Van, T. M. & Blank, C. U. A user’s perspective on GeoMxTM digital spatial profiling. Immunooncol. Technol. 1, 11–18 (2019).
Acknowledgements
The work of S.H.G. is supported by a Kay Kendall Leukaemia Fund Fellowship. The work of J.B.I. is supported by the NCI (K12CA090354) and Conquer Cancer Foundation-Sontag Foundation Young Investigator Award. The work of D.A.B. is supported by the DF/HCC Kidney Cancer SPORE Career Enhancement Program (P50CA101942-15), DOD CDMRP (KC170216, KC190130) and the DOD Academy of Kidney Cancer Investigators (KC190128). The work of D.B.K. is supported by the NIH/NCI (R21 CA216772-01A1 and NCI-SPORE-2P50CA101942-11A1). The work of K.J.L. is supported by the NIH/NCI (U24 CA224331 R01CA229261-01, NIH/NCI P01CA229092) and the NIH/NIAID (U19 AI082630).
Author information
Authors and Affiliations
Contributions
All authors researched data for this manuscript. D.B.K., S.H.G., J.B.I. and D.A.B. made substantial contributions to discussions of content. All authors wrote the manuscript and reviewed and/or edited the manuscript prior to submission.
Corresponding author
Ethics declarations
Competing interests
D.A.B. has received non-financial support from Bristol Myers Squibb, honoraria from LM Education/Exchange Services and personal fees from Adept Field Solutions, Blueprint Partnerships, Charles River Associates, Dedham Group, Defined Health, Insight Strategy, Octane Global, Slingshot Insights and Trinity Group. D.B.K. has acted as an advisor of and has received consulting fees from Neon Therapeutics and owns equity in Aduro Biotech, Agenus, Armata Pharmaceuticals, Breakbio, BioMarin Pharmaceutical, Bristol Myers Squibb, Celldex Therapeutics, Editas Medicine, Exelixis, Gilead Sciences, IMV, Lexicon Pharmaceuticals, Moderna, Regeneron Pharmaceuticals and Stemline Therapeutics. The other authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Gohil, S.H., Iorgulescu, J.B., Braun, D.A. et al. Applying high-dimensional single-cell technologies to the analysis of cancer immunotherapy. Nat Rev Clin Oncol 18, 244–256 (2021). https://doi.org/10.1038/s41571-020-00449-x
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41571-020-00449-x
This article is cited by
-
T-cell infiltration and its regulatory mechanisms in cancers: insights at single-cell resolution
Journal of Experimental & Clinical Cancer Research (2024)
-
Translational research on drug development and biomarker discovery for hepatocellular carcinoma
Journal of Biomedical Science (2024)
-
Semi-automated approaches for interrogating spatial heterogeneity of tissue samples
Scientific Reports (2024)
-
cnnImpute: missing value recovery for single cell RNA sequencing data
Scientific Reports (2024)
-
Computational immunogenomic approaches to predict response to cancer immunotherapies
Nature Reviews Clinical Oncology (2024)