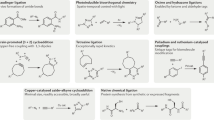
Abstract
Bioorthogonal chemistries enable researchers to interrogate biomolecules in living systems. These reactions are highly selective and biocompatible, and can be performed in many complex environments. However, like any organic transformation, there is no perfect bioorthogonal reaction. Choosing the ‘best fit’ for a desired application is crucial. Correspondingly, there must be a variety of chemistries — spanning a range of rates and other features — to choose from. Over the past few years, considerable strides have been made towards not only expanding the number of bioorthogonal chemistries but also fine-tuning existing reactions for particular applications. In this Review, we highlight recent advances in the development of bioorthogonal reactions, focusing on how principles of physical organic chemistry have guided probe design. The continued expansion of this toolset will provide precisely tuned reagents for manipulating bonds in distinct environments.

This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
References
Rodriguez, E. A. et al. The growing and glowing toolbox of fluorescent and photoactive proteins. Trends Biochem. Sci. 42, 111–129 (2017).
Row, R. D. & Prescher, J. A. Constructing new bioorthogonal reagents and reactions. Acc. Chem. Res. 51, 1073–1081 (2018).
Tian, Y. & Lin, Q. Fitness factors for bioorthogonal chemical probes. ACS Chem. Biol. 14, 2489–2496 (2019).
Sletten, E. M. & Bertozzi, C. R. Bioorthogonal chemistry: fishing for selectivity in a sea of functionality. Angew. Chem. Int. Ed. 48, 6974–6998 (2009).
Lang, K. & Chin, J. W. Cellular incorporation of unnatural amino acids and bioorthogonal labeling of proteins. Chem. Rev. 114, 4764–4806 (2014).
Carrell, T. et al. in Cycloadditions in Bioorthogonal Chemistry (eds Vrabel, M. & Carell, T.) 1–157 (Springer, 2016).
Andresen, M., Schmitz-Salue, R. & Jakobs, S. Short tetracysteine tags to β-tubulin demonstrate the significance of small labels for live cell imaging. Mol. Biol. Cell 15, 5616–5622 (2004).
Grammel, M. & Hang, H. C. Chemical reporters for biological discovery. Nat. Chem. Biol. 9, 475–484 (2013).
Chai, Q.-Y., Yang, Z., Lin, H.-W. & Han, B.-N. Alkynyl-containing peptides of marine origin: a review. Mar. Drugs 14, 216 (2016).
Jewett, J. C. & Bertozzi, C. R. Cu-free click cycloaddition reactions in chemical biology. Chem. Soc. Rev. 39, 1272–1279 (2010).
Selvaraj, R. & Fox, J. M. trans-Cyclooctene — a stable, voracious dienophile for bioorthogonal labeling. Curr. Opin. Chem. Biol. 17, 753–760 (2013).
Dommerholt, J., Rutjes, F. P. J. T. & van Delft, F. L. in Cycloadditions in Bioorthogonal Chemistry (eds Vrabel, M. & Carell, T.) 57–76 (Springer, 2016).
Kölmel, D. K. & Kool, E. T. Oximes and hydrazones in bioconjugation: mechanism and catalysis. Chem. Rev. 117, 10358–10376 (2017).
Mahal, L. K., Yarema, K. J. & Bertozzi, C. R. Engineering chemical reactivity on cell surfaces through oligosaccharide biosynthesis. Science 276, 1125–1128 (1997).
Datta, D., Wang, P., Carrico, I. S., Mayo, S. L. & Tirrell, D. A. A designed phenylalanyl-tRNA synthetase variant allows efficient in vivo incorporation of aryl ketone functionality into proteins. J. Am. Chem. Soc. 124, 5652–5653 (2002).
Rabuka, D., Rush, J. S., deHart, G. W., Wu, P. & Bertozzi, C. R. Site-specific chemical protein conjugation using genetically encoded aldehyde tags. Nat. Protoc. 7, 1052–1067 (2012).
Hardisty, R. E., Kawasaki, F., Sahakyan, A. B. & Balasubramanian, S. Selective chemical labeling of natural T modifications in DNA. J. Am. Chem. Soc. 137, 9270–9272 (2015).
Dirksen, A., Dirksen, S., Hackeng, T. M. & Dawson, P. E. Nucleophilic catalysis of hydrazone formation and transimination: implications for dynamic covalent chemistry. J. Am. Chem. Soc. 128, 15602–15603 (2006).
Dirksen, A., Hackeng, T. M. & Dawson, P. E. Nucleophilic catalysis of oxime ligation. Angew. Chem. Int. Ed. 45, 7581–7584 (2006).
Dirksen, A. & Dawson, P. E. Rapid oxime and hydrazone ligations with aromatic aldehydes for biomolecular labeling. Bioconjug. Chem. 19, 2543–2548 (2008).
Larsen, D. et al. Exceptionally rapid oxime and hydrazone formation promoted by catalytic amine buffers with low toxicity. Chem. Sci. 9, 5252–5259 (2018).
Saxon, E. & Bertozzi, C. R. Cell surface engineering by a modified Staudinger reaction. Science 287, 2007–2010 (2000).
Kiick, K. L., Saxon, E., Tirrell, D. A. & Bertozzi, C. R. Incorporation of azides into recombinant proteins for chemoselective modification by the Staudinger ligation. Proc. Natl Acad. Sci. USA 99, 19–24 (2002).
Saxon, E. et al. Investigating cellular metabolism of synthetic azidosugars with the Staudinger ligation. J. Am. Chem. Soc. 124, 14893–14902 (2002).
Cai, J., Li, X., Yue, X. & Taylor, J. S. Nucleic acid-triggered fluorescent probe activation by the Staudinger reaction. J. Am. Chem. Soc. 126, 16324–16325 (2004).
Hannoush, R. N. & Sun, J. The chemical toolbox for monitoring protein fatty acylation and prenylation. Nat. Chem. Biol. 6, 498–506 (2010).
Chuh, K. N. & Pratt, M. R. Chemical methods for the proteome-wide identification of posttranslationally modified proteins. Curr. Opin. Chem. Biol. 24, 27–37 (2015).
Prescher, J. A., Dube, D. H. & Bertozzi, C. R. Chemical remodelling of cell surfaces in living animals. Nature 430, 873–877 (2004).
Meldal, M. & Tornøe, C. W. Cu-catalyzed azide–alkyne cycloaddition. Chem. Rev. 108, 2952–3015 (2008).
Thirumurugan, P., Matosiuk, D. & Jozwiak, K. Click chemistry for drug development and diverse chemical–biology applications. Chem. Rev. 113, 4905–4979 (2013).
Lampkowski, J. S., Villa, J. K., Young, T. S. & Young, D. D. Development and optimization of Glaser–Hay bioconjugations. Angew. Chem. Int. Ed. 54, 9343–9346 (2015).
Gaetke, L. M. & Chow, C. K. Copper toxicity, oxidative stress, and antioxidant nutrients. Toxicology 189, 147–163 (2003).
Soares, E. V., Hebbelinck, K. & Soares, H. M. V. M. Toxic effects caused by heavy metals in the yeast Saccharomyces cerevisiae: a comparative study. Can. J. Microbiol. 49, 336–343 (2003).
Kennedy, D. C. et al. Cellular consequences of copper complexes used to catalyze bioorthogonal click reactions. J. Am. Chem. Soc. 133, 17993–18001 (2011).
del Amo, D. S. et al. Biocompatible copper(I) catalysts for in vivo imaging of glycans. J. Am. Chem. Soc. 132, 16893–16899 (2010).
Besanceney-Webler, C. et al. Increasing the efficacy of bioorthogonal click reactions for bioconjugation: a comparative study. Angew. Chem. Int. Ed. 50, 8051–8056 (2011).
Uttamapinant, C. et al. Fast, cell-compatible click chemistry with copper-chelating azides for biomolecular labeling. Angew. Chem. Int. Ed. 51, 5852–5856 (2012).
Yang, M. et al. Biocompatible click chemistry enabled compartment-specific pH measurement inside E. coli. Nat. Commun. 5, 4981 (2014).
Wittig, G. & Krebs, A. Zur Existenz niedergliedriger cycloalkine, I [German]. Chem. Ber. 94, 3260–3275 (1961).
Agard, N. J., Prescher, J. A. & Bertozzi, C. R. A strain-promoted [3+2] azide–alkyne cycloaddition for covalent modification of biomolecules in living systems. J. Am. Chem. Soc. 126, 15046–15047 (2004).
Rydahl, M. G., Hansen, A. R., Kračun, S. K. & Mravec, J. Report on the current inventory of the toolbox for plant cell wall analysis: proteinaceous and small molecular probes. Front. Plant Sci. 9, 581 (2018).
Yuet, K. P. et al. Cell-specific proteomic analysis in Caenorhabditis elegans. Proc. Natl Acad. Sci. USA 112, 2705–2710 (2015).
tom Dieck, S. et al. Direct visualization of newly synthesized target proteins in situ. Nat. Methods 12, 411–414 (2015).
Ernst, R. J. et al. Genetic code expansion in the mouse brain. Nat. Chem. Biol. 12, 776–778 (2016).
Alvarez-Castelao, B. et al. Cell-type-specific metabolic labeling of nascent proteomes in vivo. Nat. Biotechnol. 35, 1196–1201 (2017).
Krogager, T. P. et al. Labeling and identifying cell-specific proteomes in the mouse brain. Nat. Biotechnol. 36, 156–159 (2018).
van Geel, R., Prujin, G. J. M., van Delft, F. L. & Boelens, W. C. Preventing thiol-yne addition improves the specificity of strain-promoted azide–alkyne cycloaddition. Bioconjug. Chem. 23, 392–398 (2012).
Blackman, M. L., Royzen, M. & Fox, J. M. Tetrazine ligation: fast bioconjugation based on inverse-electron-demand Diels–Alder reactivity. J. Am. Chem. Soc. 130, 13518–13519 (2008).
Taylor, M. T., Blackman, M. L., Dmitrenko, O. & Fox, J. M. Design and synthesis of highly reactive dienophiles for the tetrazine–trans-cyclooctene ligation. J. Am. Chem. Soc. 133, 9646–9649 (2011).
Devaraj, N. K., Upadhyay, R., Haun, J. B., Hilderbrand, S. A. & Weissleder, R. Fast and sensitive pretargeted labeling of cancer cells through a tetrazine/trans-cyclooctene cycloaddition. Angew. Chem. Int. Ed. 48, 7013–7016 (2009).
Wang, M. et al. Conformationally strained trans-cyclooctene (sTCO) enables the rapid construction of 18F-PET probes via tetrazine ligation. Theranostics 6, 887–895 (2016).
Billaud, E. M. F. et al. Pretargeted PET imaging using a bioorthogonal 18F-labeled trans-cyclooctene in an ovarian carcinoma model. Bioconjug. Chem. 28, 2915–2920 (2017).
Rossin, R. et al. Highly reactive trans-cyclooctene tags with improved stability for Diels–Alder chemistry in living systems. Bioconjug. Chem. 24, 1210–1217 (2013).
Läppchen, T. et al. DOTA-tetrazine probes with modified linkers for tumor pretargeting. Nucl. Med. Biol. 55, 19–26 (2017).
Bilton, H. A., Ahmad, Z., Janzen, N., Czorny, S. & Valliant, J. F. Preparation and evaluation of 99mTc-labeled tridentate chelates for pre-targeting using bioorthogonal chemistry. J. Vis. Exp. 120, e55188 (2017).
Versteegen, R. M., Rossin, R., ten Hoeve, W., Janssen, H. M. & Robillard, M. S. Click to release: instantaneous doxorubicin elimination upon tetrazine ligation. Angew. Chem. Int. Ed. 52, 14112–14116 (2013).
Rossin, R. et al. Chemically triggered drug release from an antibody–drug conjugate leads to potent antitumour activity in mice. Nat. Commun. 9, 1484 (2018).
Versteegen, R. M. et al. Click-to-release from trans-cyclooctenes: mechanistic insights and expansion of scope from established carbamate to remarkable ether cleavage. Angew. Chem. Int. Ed. 57, 10494–10499 (2018).
Mejia Oneto, J. M., Khan, I., Seebald, L. & Royzen, M. In vivo bioorthogonal chemistry enables local hydrogel and systemic pro-drug to treat soft tissue sarcoma. ACS Cent. Sci. 2, 476–482 (2016).
Zhang, G. et al. Bioorthogonal chemical activation of kinases in living systems. ACS Cent. Sci. 2, 325–331 (2016).
van der Gracht, A. M. F. et al. Chemical control over T-cell activation in vivo using deprotection of trans-cyclooctene-modified epitopes. ACS Chem. Biol. 13, 1569–1576 (2018).
Fan, X. et al. Optimized tetrazine derivatives for rapid bioorthogonal decaging in living cells. Angew. Chem. Int. Ed. 55, 14046–14050 (2016).
Schmidt, P., Zhou, L., Tishinov, K., Zimmermann, K. & Gillingham, D. Dialdehydes lead to exceptionally fast bioconjugations at neutral pH by virtue of a cyclic intermediate. Angew. Chem. Int. Ed. 53, 10928–10931 (2014).
Dilek, O., Lei, Z., Mukherjee, K. & Bane, S. Rapid formation of a stable boron–nitrogen heterocycle in dilute, neutral aqueous solution for bioorthogonal coupling reactions. Chem. Commun. 51, 16992–16995 (2015).
Schmidt, P., Stress, C. & Gillingham, D. Boronic acids facilitate rapid oxime condensations at neutral pH. Chem. Sci. 6, 3329–3333 (2015).
Meadows, M. K., Roesner, E. K., Lynch, V. M., James, T. D. & Anslyn, E. V. Boronic acid mediated coupling of catechols and N-hydroxylamines: a bioorthogonal reaction to label peptides. Org. Lett. 19, 3179–3182 (2017).
Bandyopadhyay, A., Cambray, S. & Gao, J. Fast diazaborine formation of semicarbazide enables facile labeling of bacterial pathogens. J. Am. Chem. Soc. 139, 871–878 (2017).
Akgun, B. et al. Synergic “click” boronate/thiosemicarbazone system for fast and irreversible bioorthogonal conjugation in live cells. J. Am. Chem. Soc. 139, 14285–14291 (2017).
Gu, H. et al. Formation of hydrazones and stabilized boron–nitrogen heterocycles in aqueous solution from carbohydrazides and ortho-formylphenylboronic acids. Org. Biomol. Chem. 15, 7543–7548 (2017).
Bandyopadhyay, A., Cambray, S. & Gao, J. Fast and selective labeling of N-terminal cysteines at neutral pH via thiazolidino boronate formation. Chem. Sci. 7, 4589–4593 (2016).
Faustino, H., Silva, M. J. S. A., Veiros, L. F., Bernardes, G. J. L. & Gois, P. M. P. Iminoboronates are efficient intermediates for selective, rapid and reversible N-terminal cysteine functionalisation. Chem. Sci. 7, 5052–5058 (2016).
Sundhoro, M., Jeon, S., Park, J., Ramström, O. & Yan, M. Perfluoroaryl azide Staudinger reaction: a fast and bioorthogonal reaction. Angew. Chem. Int. Ed. 56, 12117–12121 (2017).
Meguro, T. et al. Staudinger reaction using 2,6-dichlorophenyl azide derivatives for robust aza-ylide formation applicable to bioconjugation in living cells. Chem. Commun. 54, 7904–7907 (2018).
Saneyoshi, H. et al. Triphenylphosphinecarboxamide: an effective reagent for the reduction of azides and its application to nucleic acid detection. Org. Lett. 16, 30–33 (2014).
Luo, J., Liu, Q., Morihiro, K. & Deiters, A. Small-molecule control of protein function through Staudinger reduction. Nat. Chem. 8, 1027–1034 (2016).
Lukasak, B., Morihiro, K. & Deiters, A. Aryl azides as phosphine-activated switches for small molecule function. Sci. Rep. 9, 1470 (2019).
Ren, G., Zheng, Q. & Wang, H. Aryl fluorosulfate trapped Staudinger reduction. Org. Lett. 19, 1582–1585 (2017).
Böhrsch, V., Serwa, R., Majkut, P., Krause, E. & Hackenberger, C. P. R. Site-specific functionalisation of proteins by a Staudinger-type reaction using unsymmetrical phosphites. Chem. Commun. 46, 3176–3178 (2010).
Serwa, R. et al. Site-specific PEGylation of proteins by a Staudinger-phosphite reaction. Chem. Sci. 1, 596–602 (2010).
Nischan, N., Kasper, M.-A., Mathew, T. & Hackenberger, C. P. R. Bis(arylmethyl)-substituted unsymmetrical phosphites for the synthesis of lipidated peptides via Staudinger-phosphite reactions. Org. Biomol. Chem. 14, 7500–7508 (2016).
Vallée, M. R. J. et al. Staudinger-phosphonite reactions for the chemoselective transformation of azido-containing peptides and proteins. Org. Lett. 13, 5440–5443 (2011).
Kasper, M.-A. et al. Vinylphosphonites for Staudinger-induced chemoselective peptide cyclization and functionalization. Chem. Sci. 10, 6322–6329 (2019).
Kasper, M.-A. et al. Cysteine-selective phosphonamidate electrophiles for modular protein bioconjugations. Angew. Chem. Int. Ed. 58, 11625–11630 (2019).
Lee, Y.-J., Kurra, Y. & Liu, W. R. Phospha-Michael addition as a new click reaction for protein functionalization. ChemBioChem 17, 456–461 (2016).
Bos, J. & Muir, T. W. A chemical probe for protein crotonylation. J. Am. Chem. Soc. 140, 4757–4760 (2018).
Shih, H.-W. & Prescher, J. A. A bioorthogonal ligation of cyclopropenones mediated by triarylphosphines. J. Am. Chem. Soc. 137, 10036–10039 (2015).
Row, R. D., Shih, H.-W., Alexander, A. T., Mehl, R. A. & Prescher, J. A. Cyclopropenones for metabolic targeting and sequential bioorthogonal labeling. J. Am. Chem. Soc. 139, 7370–7375 (2017).
Row, R. D. & Prescher, J. A. A cyclopropenethione-phosphine ligation for rapid biomolecule labeling. Org. Lett. 20, 5614–5617 (2018).
Heiss, T. K. & Prescher, J. A. Cyclopropeniminium ions exhibit unique reactivity profiles with bioorthogonal phosphines. J. Org. Chem. 84, 7443–7448 (2019).
Newberry, R. W., VanVeller, B., Guzei, I. A. & Raines, R. T. n→π* interactions of amides and thioamides: implications for protein stability. J. Am. Chem. Soc. 135, 7843–7846 (2013).
Walters, C. R. et al. The effects of thioamide backbone substitution on protein stability: a study in α-helical, β-sheet, and polyproline II helical contexts. Chem. Sci. 8, 2868–2877 (2017).
McKay, C. S., Moran, J. & Pezacki, J. P. Nitrones as dipoles for rapid strain-promoted 1,3-dipolar cycloadditions with cyclooctynes. Chem. Commun. 46, 931–933 (2010).
McKay, C. S., Chigrinova, M., Blake, J. A. & Pezacki, J. P. Kinetics studies of rapid strain-promoted [3+2]-cycloadditions of nitrones with biaryl-aza-cyclooctynone. Org. Biomol. Chem. 10, 3066–3070 (2012).
MacKenzie, D. A. & Pezacki, J. P. Kinetics studies of rapid strain-promoted [3+2] cycloadditions of nitrones with bicyclo[6.1.0]nonyne. Can. J. Chem. 92, 337–340 (2014).
Gunawardene, P. N., Luo, W., Polgar, A. M., Corrigan, J. F. & Workentin, M. S. Highly electron-deficient pyridinium-nitrones for rapid and tunable inverse-electron-demand strain-promoted alkyne-nitrone cycloaddition. Org. Lett. 21, 5547–5551 (2019).
McKay, C. S., Blake, J. A., Cheng, J., Danielson, D. C. & Pezacki, J. P. Strain-promoted cycloadditions of cyclic nitrones with cyclooctynes for labeling human cancer cells. Chem. Commun. 47, 10040–10042 (2011).
Sherratt, A. R. et al. Copper-catalysed cycloaddition reactions of nitrones and alkynes for bioorthogonal labelling of living cells. RSC Adv. 4, 46966–46969 (2014).
Sherratt, A. R. et al. Dual strain-promoted alkyne–nitrone cycloadditions for simultaneous labeling of bacterial peptidoglycans. Bioconjug. Chem. 27, 1222–1226 (2016).
Decuypère, E., Plougastel, L., Audisio, D. & Taran, F. Sydnone–alkyne cycloaddition: applications in synthesis and bioconjugation. Chem. Commun. 53, 11515–11527 (2017).
Kolodych, S. et al. Discovery of chemoselective and biocompatible reactions using a high-throughput immunoassay screening. Angew. Chem. Int. Ed. 52, 12056–12060 (2013).
Wallace, S. & Chin, J. W. Strain-promoted sydnone bicyclo-[6.1.0]-nonyne cycloaddition. Chem. Sci. 5, 1742–1744 (2014).
Plougastel, L. et al. 4-Halogeno-sydnones for fast strain promoted cycloaddition with bicyclo-[6.1.0]-nonyne. Chem. Commun. 50, 9376–9378 (2014).
Liu, H. et al. Ultrafast click chemistry with fluorosydnones. Angew. Chem. Int. Ed. 55, 12073–12077 (2016).
Mix, K. A., Aronoff, M. R. & Raines, R. T. Diazo compounds: versatile tools for chemical biology. ACS Chem. Biol. 11, 3233–3244 (2016).
Moran, J., McKay, C. S. & Pezacki, J. P. Strain-promoted 1,3-dipolar cycloadditions of diazo compounds with cyclooctynes. Can. J. Chem. 89, 148–151 (2011).
Sanders, B. C. et al. Metal-free sequential [3+ 2]-dipolar cycloadditions using cyclooctynes and 1,3-dipoles of different reactivity. J. Am. Chem. Soc. 133, 949–957 (2011).
Josa-Culleré, L., Wainman, Y. A., Brindle, K. M. & Leeper, F. J. Diazo group as a new chemical reporter for bioorthogonal labelling of biomolecules. RSC Adv. 4, 52241–52244 (2014).
Anderson, K. A., Aronoff, M. R., McGrath, N. A. & Raines, R. T. Diazo groups endure metabolism and enable chemoselectivity in cellulo. J. Am. Chem. Soc. 137, 2412–2415 (2015).
McGrath, N. A. & Raines, R. T. Diazo compounds as highly tunable reactants in 1,3-dipolar cycloaddition reactions with cycloalkynes. Chem. Sci. 3, 3237–3240 (2012).
Aronoff, M. R., Gold, B. & Raines, R. T. 1,3-Dipolar cycloadditions of diazo compounds in the presence of azides. Org. Lett. 18, 1538–1541 (2016).
Lim, R. K. V. & Lin, Q. Azirine ligation: fast and selective protein conjugation via photoinduced azirine–alkene cycloaddition. Chem. Commun. 46, 7993–7995 (2010).
King, M. & Wagner, A. Developments in the field of bioorthogonal bond forming reactions — past and present trends. Bioconjug. Chem. 25, 825–839 (2014).
Schäfer, R. J. B. et al. The bioorthogonal isonitrile–chlorooxime ligation. J. Am. Chem. Soc. 141, 18644–18648 (2019).
Yu, Z., Lim, R. K. V. & Lin, Q. Synthesis of macrocyclic tetrazoles for rapid photoinduced bioorthogonal 1,3-dipolar cycloaddition reactions. Chem. Eur. J. 16, 13325–13329 (2010).
Yu, Z., Pan, Y., Wang, Z., Wang, J. & Lin, Q. Genetically encoded cyclopropene directs rapid, photoclick-chemistry-mediated protein labeling in mammalian cells. Angew. Chem. Int. Ed. 51, 10600–10604 (2012).
Kamber, D. N. et al. Isomeric cyclopropenes exhibit unique bioorthogonal reactivities. J. Am. Chem. Soc. 135, 13680–13683 (2013).
An, P., Yu, Z. & Lin, Q. Design of oligothiophene-based tetrazoles for laser-triggered photoclick chemistry in living cells. Chem. Commun. 49, 9920–9922 (2013).
Yu, Z., Ohulchanskyy, T. Y., An, P., Prasad, P. N. & Lin, Q. Fluorogenic, two-photon-triggered photoclick chemistry in live mammalian cells. J. Am. Chem. Soc. 135, 16766–16769 (2013).
Tasdelen, M. A. & Yagci, Y. Light-induced click reactions. Angew. Chem. Int. Ed. 52, 5930–5938 (2013).
Li, J. et al. Visible light-initiated bioorthogonal photoclick cycloaddition. J. Am. Chem. Soc. 140, 14542–14546 (2018).
Zhang, X. et al. Photo-accelerated “click” reaction between diarylsydnones and ring-strained alkynes for bioorthogonal ligation. Chem. Commun. 55, 7187–7190 (2019).
Lim, R. K. V. & Lin, Q. Photoinducible bioorthogonal chemistry: a spatiotemporally controllable tool to visualize and perturb proteins in live cells. Acc. Chem. Res. 44, 828–839 (2011).
An, P., Lewandowski, T. M., Erbay, T. G., Liu, P. & Lin, Q. Sterically shielded, stabilized nitrile imine for rapid bioorthogonal protein labeling in live cells. J. Am. Chem. Soc. 140, 4860–4868 (2018).
de Almeida, G., Sletten, E. M., Nakamura, H., Palaniappan, K. K. & Bertozzi, C. R. Thiacycloalkynes for copper-free click chemistry. Angew. Chem. Int. Ed. 51, 2443–2447 (2012).
King, M., Baati, R. & Wagner, A. New tetramethylthiepinium (TMTI) for copper-free click chemistry. Chem. Commun. 48, 9308–9309 (2012).
de Almeida, G., Townsend, L. C. & Bertozzi, C. R. Synthesis and reactivity of dibenzoselenacycloheptynes. Org. Lett. 15, 3038–3041 (2013).
Ning, X., Guo, J., Wolfert, M. A. & Boons, G.-J. Visualizing metabolically labeled glycoconjugates of living cells by copper-free and fast Huisgen cycloadditions. Angew. Chem. Int. Ed. 47, 2253–2255 (2008).
Stöckmann, H. et al. Development and evaluation of new cyclooctynes for cell surface glycan imaging in cancer cells. Chem. Sci. 2, 932–936 (2011).
Friscourt, F. et al. Polar dibenzocyclooctynes for selective labeling of extracellular glycoconjugates of living cells. J. Am. Chem. Soc. 134, 5381–5389 (2012).
Varga, B. R., Kállay, M., Hegyi, K., Béni, S. & Kele, P. A non-fluorinated monobenzocyclooctyne for rapid copper-free click reactions. Chem. Eur. J. 18, 822–828 (2012).
Dommerholt, J. et al. Readily accessible bicyclononynes for bioorthogonal labeling and three-dimensional imaging of living cells. Angew. Chem. Int. Ed. 49, 9422–9425 (2010).
Agard, N. J., Baskin, J. M., Prescher, J. A., Lo, A. & Bertozzi, C. R. A comparative study of bioorthogonal reactions with azides. ACS Chem. Biol. 1, 644–648 (2006).
Baskin, J. M. et al. Copper-free click chemistry for dynamic in vivo imaging. Proc. Natl Acad. Sci. USA 104, 16793–16797 (2007).
Sletten, E. M., Nakamura, H., Jewett, J. C. & Bertozzi, C. R. Difluorobenzocyclooctyne: synthesis, reactivity, and stabilization by β-cyclodextrin. J. Am. Chem. Soc. 132, 11799–11805 (2010).
Li, W. et al. Fluorodibenzocyclooctynes: a trackable click reagent with enhanced reactivity. Chem. Eur. J. 25, 10328–10332 (2019).
Sletten, E. M. & Bertozzi, C. R. A hydrophilic azacyclooctyne for Cu-free click chemistry. Org. Lett. 10, 3097–3099 (2008).
Jewett, J. C., Sletten, E. M. & Bertozzi, C. R. Rapid Cu-free click chemistry with readily synthesized biarylazacyclooctynones. J. Am. Chem. Soc. 132, 3688–3690 (2010).
Debets, M. F. et al. Aza-dibenzocyclooctynes for fast and efficient enzyme PEGylation via copper-free (3+2) cycloaddition. Chem. Commun. 46, 97–99 (2010).
Gold, B., Shevchenko, N. E., Bonus, N., Dudley, G. B. & Alabugin, I. V. Selective transition state stabilization via hyperconjugative and conjugative assistance: stereoelectronic concept for copper-free click chemistry. J. Org. Chem. 77, 75–89 (2012).
Gold, B., Batsomboon, P., Dudley, G. B. & Alabugin, I. V. Alkynyl crown ethers as a scaffold for hyperconjugative assistance in noncatalyzed azide–alkyne click reactions: ion sensing through enhanced transition-state stabilization. J. Org. Chem. 79, 6221–6232 (2014).
Kaneda, K., Naruse, R. & Yamamoto, S. 2-Aminobenzenesulfonamide-containing cyclononyne as adjustable click reagent for strain-promoted azide–alkyne cycloaddition. Org. Lett. 19, 1096–1099 (2017).
Burke, E. G., Gold, B., Hoang, T. T., Raines, R. T. & Schomaker, J. M. Fine-tuning strain and electronic activation of strain-promoted 1,3-dipolar cycloadditions with endocyclic sulfamates in SNO-OCTs. J. Am. Chem. Soc. 139, 8029–8037 (2017).
Ni, R., Mitsuda, N., Kashiwagi, T., Igawa, K. & Tomooka, K. Heteroatom-embedded medium-sized cycloalkynes: concise synthesis, structural analysis, and reactions. Angew. Chem. Int. Ed. 54, 1190–1194 (2015).
Del Grosso, A. et al. Strained alkynes derived from 2,2′-dihydroxy-1,1′-biaryls; synthesis and copper-free cycloaddition with azides. Org. Biomol. Chem. 15, 4517–4521 (2017).
Mistry, A. et al. Synthesis and cycloaddition reactions of strained alkynes derived from 2,2′-dihydroxy-1,1′-biaryls. Org. Biomol. Chem. 16, 8965–8975 (2018).
Harris, T. et al. Twisted cycloalkynes and remote activation of “click” reactivity. Chem 3, 629–640 (2017).
Meier, H., Schuh-Popitz, C. & Peiersen, H. Isolation of a highly strained bicyclic alkyne. Angew. Chem. Int. Ed. Engl. 20, 270–271 (1981).
Ho, S. H. & Tirrell, D. A. Enzymatic labeling of bacterial proteins for super-resolution imaging in live cells. ACS Cent. Sci. 5, 1911–1919 (2019).
Liu, F., Liang, Y. & Houk, K. N. Bioorthogonal cycloadditions: computational analysis with the distortion/interaction model and predictions of reactivities. Acc. Chem. Res. 50, 2297–2308 (2017).
Bickelhaupt, F. M. & Houk, K. N. Analyzing reaction rates with the distortion/interaction-activation strain model. Angew. Chem. Int. Ed. 56, 10070–10086 (2017).
Gordon, C. G. et al. Reactivity of biarylazacyclooctynones in copper-free click chemistry. J. Am. Chem. Soc. 134, 9199–9208 (2012).
Tao, H. et al. Origins of halogen effects in bioorthogonal sydnone cycloadditions. Chem. Commun. 54, 5082–5085 (2018).
Liu, F., Paton, R. S., Kim, S., Liang, Y. & Houk, K. N. Diels–Alder reactivities of strained and unstrained cycloalkenes with normal and inverse-electron-demand dienes: activation barriers and distortion/interaction analysis. J. Am. Chem. Soc. 135, 15642–15649 (2013).
Liu, F., Liang, Y. & Houk, K. N. Theoretical elucidation of the origins of substituent and strain effects on the rates of Diels–Alder reactions of 1,2,4,5-tetrazines. J. Am. Chem. Soc. 136, 11483–11493 (2014).
Darko, A. et al. Conformationally strained trans-cyclooctene with improved stability and excellent reactivity in tetrazine ligation. Chem. Sci. 5, 3770–3776 (2014).
Kozma, E. et al. Hydrophilic trans-cyclooctenylated noncanonical amino acids for fast intracellular protein labeling. ChemBioChem 17, 1518–1524 (2016).
Knorr, G., Kozma, E., Herner, A., Lemke, E. A. & Kele, P. New red-emitting tetrazine-phenoxazine fluorogenic labels for live-cell intracellular bioorthogonal labeling schemes. Chem. Eur. J. 22, 8972–8979 (2016).
Lambert, W. D. et al. Computationally guided discovery of a reactive, hydrophilic trans-5-oxocene dienophile for bioorthogonal labeling. Org. Biomol. Chem. 15, 6640–6644 (2017).
Siegl, S. J. et al. Design and synthesis of aza-bicyclononene dienophiles for rapid fluorogenic ligations. Chem. Eur. J. 24, 2426–2432 (2018).
Bach, R. D. Ring strain energy in the cyclooctyl system. The effect of strain energy on [3+2] cycloaddition reactions with azides. J. Am. Chem. Soc. 131, 5233–5243 (2009).
Santucci, J. III, Sanzone, J. R. & Woerpel, K. A. [4+2] cycloadditions of seven- membered-ring trans-alkenes: decreasing reactivity with increasing substitution of the seven-membered ring. Eur. J. Org. Chem. 2016, 2933–2943 (2016).
Fang, Y. et al. Photochemical syntheses, transformations, and bioorthogonal chemistry of trans-cycloheptene and sila trans-cycloheptene Ag(I) complexes. Chem. Sci. 9, 1953–1963 (2018).
Patterson, D. M., Nazarova, L. A., Xie, B., Kamber, D. N. & Prescher, J. A. Functionalized cyclopropenes as bioorthogonal chemical reporters. J. Am. Chem. Soc. 134, 18638–18643 (2012).
Yang, J., Šečkutė, J., Cole, C. M. & Devaraj, N. K. Live-cell imaging of cyclopropene tags with fluorogenic tetrazine cycloadditions. Angew. Chem. Int. Ed. 51, 7476–7479 (2012).
Ravasco, J. M. J. M., Monteiro, C. M. & Trindade, A. F. Cyclopropenes: a new tool for the study of biological systems. Org. Chem. Front. 4, 1167–1198 (2017).
Elliot, T. S. et al. Proteome labeling and protein identification in specific tissues and at specific developmental stages in an animal. Nat. Biotechnol. 32, 465–472 (2014).
Yu, Z. & Lin, Q. Design of spiro[2.3]hex-1-ene, a genetically encodable double-strained alkene for superfast photoclick chemistry. J. Am. Chem. Soc. 136, 4153–4156 (2014).
Ramil, C. P. et al. Spirohexene-tetrazine ligation enables bioorthogonal labeling of class B G protein-coupled receptors in live cells. J. Am. Chem. Soc. 139, 13376–13386 (2017).
An, P., Wu, H.-Y., Lewandowski, T. M. & Lin, Q. Hydrophilic azaspiroalkenes as robust bioorthogonal reporters. Chem. Commun. 54, 14005–14008 (2018).
Kumar, P., Jiang, T., Li, S., Zainul, O. & Laughlin, S. T. Caged cyclopropenes for controlling bioorthogonal reactivity. Org. Biomol. Chem. 16, 4081–4085 (2018).
Kumar, P. et al. Caged cyclopropenes with improved tetrazine ligation kinetics. Org. Lett. 21, 3721–3725 (2019).
Tu, J., Xu, M., Parvez, S., Peterson, R. T. & Franzini, R. M. Bioorthogonal removal of 3-isocyanopropyl groups enables the controlled release of fluorophores and drugs in vivo. J. Am. Chem. Soc. 140, 8410–8414 (2018).
Tu, J. et al. Isonitrile-responsive and bioorthogonally removable tetrazine protecting groups. Chem. Sci. 11, 169–179 (2020).
Xu, M., Deb, T., Tu, J. & Franzini, R. M. Tuning isonitrile/tetrazine chemistry for accelerated deprotection and formation of stable conjugates. J. Org. Chem. 84, 15520–15529 (2019).
Stöckmann, H., Neves, A. A., Stairs, S., Brindle, K. M. & Leeper, F. J. Exploring isonitrile-based click chemistry for ligation with biomolecules. Org. Biomol. Chem. 9, 7303–7305 (2011).
Chen, Y. et al. Addition of Isocyanide-containing amino acids to the genetic code for protein labeling and activation. ACS Chem. Biol. 14, 2793–2799 (2019).
Stairs, S. et al. Metabolic glycan imaging by isonitrile–tetrazine click chemistry. ChemBioChem 14, 1063–1067 (2013).
Niederwieser, A. et al. Two-color glycan labeling of live cells by a combination of Diels–Alder and click chemistry. Angew. Chem. Int. Ed. 52, 4265–4268 (2013).
Lee, Y.-J. et al. Genetically encoded unstrained olefins for live cell labeling with tetrazine dyes. Chem. Commun. 50, 13085–13088 (2014).
Eising, S., Lelivelt, F. & Bonger, K. M. Vinylboronic acids as fast reacting, synthetically accessible, and stable bioorthogonal reactants in the Carboni–Lindsey reaction. Angew. Chem. Int. Ed. 55, 12243–12247 (2016).
Eising, S. et al. Coordination-assisted bioorthogonal chemistry: orthogonal tetrazine ligation with vinylboronic acid and a strained alkene. ChemBioChem 19, 1648–1652 (2018).
Eising, S., Engwerda, A. H. J., Riedijk, X., Bickelhaupt, F. M. & Bonger, K. M. Highly stable and selective tetrazines for the coordination-assisted bioorthogonal ligation with vinylboronic acids. Bioconjug. Chem. 29, 3054–3059 (2018).
Eising, S., van der Linden, N. G. A., Kleinpenning, F. & Bonger, K. M. Vinylboronic acids as efficient bioorthogonal reactants for tetrazine labeling in living cells. Bioconjug. Chem. 29, 982–986 (2018).
Engelsma, S. B. et al. Acylazetine as a dienophile in bioorthogonal inverse electron-demand Diels–Alder ligation. Org. Lett. 16, 2744–2747 (2014).
de Montes, E. G. et al. Azabicyclic vinyl sulfones for residue-specific dual protein labelling. Chem. Sci. 10, 4515–4522 (2019).
Oliveira, B. L. et al. A minimal, unstrained S-allyl handle for pre-targeting Diels–Alder bioorthogonal labeling in live cells. Angew. Chem. Int. Ed. 55, 14683–14687 (2016).
Oliveira, B. L., Guo, Z. & Bernardes, G. J. L. Inverse electron demand Diels–Alder reactions in chemical biology. Chem. Soc. Rev. 46, 4895–4950 (2017).
Chen, W., Wang, D., Dai, C., Hamelberg, D. & Wang, B. Clicking 1,2,4,5-tetrazine and cyclooctynes with tunable reaction rates. Chem. Commun. 48, 1736–1738 (2012).
Wang, D. et al. 3,6-Substituted-1,2,4,5-tetrazines: tuning reaction rates for staged labeling applications. Org. Biomol. Chem. 12, 3950–3955 (2014).
Blizzard, R. J. et al. Ideal bioorthogonal reactions using a site-specifically encoded tetrazine amino acid. J. Am. Chem. Soc. 137, 10044–10047 (2015).
Lambert, W. D. et al. Installation of minimal tetrazines through silver-mediated Liebeskind–Srogl coupling with arylboronic acids. J. Am. Chem. Soc. 141, 17068–17074 (2019).
Šečkutė, J. & Devaraj, N. K. Expanding room for tetrazine ligations in the in vivo chemistry toolbox. Curr. Opin. Chem. Biol. 17, 761–767 (2013).
Zhang, H. et al. Rapid bioorthogonal chemistry turn-on through enzymatic or long wavelength photocatalytic activation of tetrazine ligation. J. Am. Chem. Soc. 138, 5978–5983 (2016).
Ehret, F., Wu, H., Alexander, S. C. & Devaraj, N. K. Electrochemical control of rapid bioorthogonal tetrazine ligations for selective functionalization of microelectrodes. J. Am. Chem. Soc. 137, 8876–8879 (2015).
Kamber, D. N. et al. 1,2,4-Triazines are versatile bioorthogonal reagents. J. Am. Chem. Soc. 137, 8388–8391 (2015).
Horner, K. A., Valette, N. M. & Webb, M. E. Strain-promoted reaction of 1,2,4-triazines with bicyclononynes. Chem. Eur. J. 21, 14376–14381 (2015).
Baalmann, M., Ziegler, M. J., Werther, P., Wilhelm, J. & Wombacher, R. Enzymatic and site-specific ligation of minimal-size tetrazines and triazines to proteins for bioconjugation and live-cell imaging. Bioconjug. Chem. 30, 1405–1414 (2019).
Peewasan, K. & Wagenknecht, H.-A. 1,2,4-Triazine-modified 2′-deoxyuridine triphosphate for efficient bioorthogonal fluorescent labeling of DNA. ChemBioChem 18, 1473–1476 (2017).
Reisacher, U. et al. Triazine-modified 7-deaza-2′-deoxyadenosines: better suited for bioorthogonal labeling of DNA by PCR than 2′-deoxyuridines. Bioconjug. Chem. 30, 1773–1780 (2019).
Siegl, S. J., Dzijak, R., Vázquez, A., Pohl, R. & Vrabel, M. The discovery of pyridinium 1,2,4-triazines with enhanced performance in bioconjugation reactions. Chem. Sci. 8, 3593–3598 (2017).
Kozhevnikov, V. N., Deary, M. E., Mantso, T., Panayiotidis, M. I. & Sims, M. T. Iridium(III) complexes of 1,2,4-triazines as potential bioorthogonal reagents: metal coordination facilitates luminogenic reaction with strained cyclooctynes. Chem. Commun. 55, 14283–14286 (2019).
Kamber, D. N. et al. Isomeric triazines exhibit unique profiles of bioorthogonal reactivity. Chem. Sci. 10, 9109–9114 (2019).
Levandowski, B. J., Abularrage, N. S., Houk, K. N. & Raines, R. T. Hyperconjugative antiaromaticity activates 4H-pyrazoles as inverse-electron-demand Diels–Alder dienes. Org. Lett. 21, 8492–8495 (2019).
Bruins, J. J., Albada, B. & van Delft, F. ortho-Quinones and analogues thereof: highly reactive intermediates for fast and selective biofunctionalization. Chem. Eur. J. 24, 4749–4756 (2018).
Borrmann, A. et al. Strain-promoted oxidation-controlled cyclooctyne–1,2-quinone cycloaddition (SPOCQ) for fast and activatable protein conjugation. Bioconjug. Chem. 26, 257–261 (2015).
Bruins, J. J. et al. Inducible, site-specific protein labeling by tyrosine oxidation–strain-promoted (4+2) cycloaddition. Bioconjug. Chem. 28, 1189–1193 (2017).
Gahtory, D., Sen, R., Kuzmyn, A. R., Escorihuela, J. & Zuilhof, H. Strain-promoted cycloaddition of cyclopropenes with o-quinones: a rapid click reaction. Angew. Chem. Int. Ed. 57, 10118–10122 (2018).
Li, Q., Dong, T., Liu, X. & Lei, X. A bioorthogonal ligation enabled by click cycloaddition of o-quinolinone quinone methide and vinyl thioether. J. Am. Chem. Soc. 135, 4996–4999 (2013).
Minard, A., Liano, D., Wang, X. & Di Antonio, M. The unexplored potential of quinone methides in chemical biology. Bioorg. Med. Chem. 27, 2298–2305 (2019).
Shih, H.-W., Kamber, D. N. & Prescher, J. A. Building better bioorthogonal reactions. Curr. Opin. Chem. Biol. 21, 103–111 (2014).
Chang, P. V., Prescher, J. A., Hangauer, M. J. & Bertozzi, C. R. Imaging cell surface glycans with bioorthogonal chemical reporters. J. Am. Chem. Soc. 129, 8400–8401 (2007).
Karver, M. R., Weissleder, R. & Hilderbrand, S. A. Bioorthogonal reaction pairs enable simultaneous, selective, multi-target imaging. Angew. Chem. Int. Ed. 51, 920–922 (2012).
Schoch, J., Staudt, M., Samanta, A., Wiessler, M. & Jäschke, A. Site-specific one-pot dual labeling of DNA by orthogonal cycloaddition chemistry. Bioconjug. Chem. 23, 1382–1386 (2012).
Hudak, J. E., Alvarez, D., Skelly, A., von Adrian, U. H. & Kasper, D. L. Illuminating vital surface molecules of symbionts in health and disease. Nat. Microbiol. 2, 17099 (2017).
Zhang, X. et al. Second generation TQ-ligation for cell organelle imaging. ACS Chem. Biol. 10, 1676–1683 (2015).
Narayanam, M. K., Liang, Y., Houk, K. N. & Murphy, J. M. Discovery of new mutually orthogonal bioorthogonal cycloaddition pairs through computational screening. Chem. Sci. 7, 1257–1261 (2016).
Shao, Z. et al. Bioorthogonal release of sulfonamides and mutually orthogonal liberation of two drugs. Chem. Commun. 54, 14089–14092 (2018).
Winz, M.-L., Linder, E. C., Becker, J. & Jäschke, A. Site-specific one-pot triple click labeling for DNA and RNA. Chem. Commun. 54, 11781–11784 (2018).
Simon, C. et al. One, two, three: a bioorthogonal triple labelling strategy for studying the dynamics of plant cell wall formation in vivo. Angew. Chem. Int. Ed. 57, 16665–16671 (2018).
Schart, V. F. et al. Triple orthogonal labeling of glycans by applying photoclick chemistry. ChemBioChem 20, 166–171 (2019).
Sletten, E. M. & Bertozzi, C. R. A bioorthogonal quadricyclane ligation. J. Am. Chem. Soc. 133, 17570–17573 (2011).
Italia, J. S. et al. Mutually orthogonal nonsense-suppression systems and conjugation chemistries for precise protein labeling at up to three distinct sites. J. Am. Chem. Soc. 141, 6204–6212 (2019).
Tu, J. et al. Stable, reactive, and orthogonal tetrazines: dispersion forces promote the cycloaddition with isonitriles. Angew. Chem. Int. Ed. 58, 9043–9048 (2019).
Kim, J. & Bertozzi, C. R. A bioorthogonal reaction of N-oxide and boron reagents. Angew. Chem. Int. Ed. 54, 15777–15781 (2015).
Dong, J., Krasnova, L., Finn, M. G. & Sharpless, K. B. Sulfur(VI) fluoride exchange (SuFEx): another good reaction for click chemistry. Angew. Chem. Int. Ed. 53, 9430–9448 (2014).
Barrow, A. S. et al. The growing applications of SuFEx click chemistry. Chem. Soc. Rev. 48, 4731–4758 (2019).
Zheng, Q. et al. SuFEx-enabled, agnostic discovery of covalent inhibitors of human neutrophil elastase. Proc. Natl Acad. Sci. USA 116, 18808–18814 (2019).
Mortenson, D. E. et al. “Inverse drug discovery” strategy to identify proteins that are targeted by latent electrophiles as exemplified by aryl fluorosulfates. J. Am. Chem. Soc. 140, 200–210 (2018).
Shannon, A. et al. Sulfonyl fluoride analogues as activity-based probes for serine proteases. ChemBioChem 13, 2327–2330 (2012).
Hett, E. C. et al. Rational targeting of active-site tyrosine residues using sulfonyl fluoride probes. ACS Chem. Biol. 10, 1094–1098 (2015).
Wang, N. et al. Genetically encoding fluorosulfate-l-tyrosine to react with lysine, histidine, and tyrosine via SuFEx in proteins in vivo. J. Am. Chem. Soc. 140, 4995–4999 (2018).
Yang, B. et al. Proximity-enhanced SuFEx chemical cross-linker for specific and multitargeting cross-linking mass spectrometry. Proc. Natl Acad. Sci. USA 115, 11162–11167 (2018).
Meng, G. et al. Modular click chemistry libraries for functional screens using a diazotizing reagent. Nature 574, 86–89 (2019).
Kabakoff, D. S., Bünzil, J. C. G., Oth, J. F. M., Hammond, W. B. & Berson, J. A. Enthalpy and kinetics of isomerization of quadricyclane to norbornadiene. Strain energy of quadricyclane. J. Am. Chem. Soc. 97, 1510–1512 (1975).
Tomlin, F. M. et al. Site-specific incorporation of quadricyclane into a protein and photocleavage of the quadricyclane ligation adduct. Bioorg. Med. Chem. 26, 5280–5290 (2018).
Martínez-Calvo, M. & Mascareñas, J. L. Organometallic catalysis in biological media and living settings. Coord. Chem. Rev. 359, 57–79 (2018).
Jang, S.-Y., Murale, D. P., Kim, A. D. & Lee, J.-S. Recent developments in metal- catalyzed bio-orthogonal reactions for biomolecule tagging. ChemBioChem 20, 1498–1507 (2019).
Vidal, C., Tomás-Gamasa, M., Destito, P., López, F. & Mascareñas, J. L. Concurrent and orthogonal gold(I) and ruthenium(II) catalysis inside living cells. Nat. Commun. 9, 1913 (2018).
Pérez-López, A. M. et al. Gold-triggered uncaging chemistry in living systems. Angew. Chem. Int. Ed. 56, 12548–12552 (2017).
Tsubokura, K. et al. In vivo gold complex catalysis within live mice. Angew. Chem. Int. Ed. 56, 3579–3584 (2017).
Chalker, J. M., Wood, C. S. C. & Davis, B. G. A convenient catalyst for aqueous and protein Suzuki–Miyaura cross-coupling. J. Am. Chem. Soc. 131, 16346–16347 (2009).
Clavadetscher, J., Indrigo, E., Chankeshwara, S. V., Lilienkampf, A. & Bradley, M. In-cell dual drug synthesis by cancer-targeting palladium catalysts. Angew. Chem. Int. Ed. 56, 6864–6868 (2017).
Li, N., Lim, R. K. V., Edwardraja, S. & Lin, Q. Copper-free Sonogashira cross-coupling for functionalization of alkyne-encoded proteins in aqueous medium and in bacterial cells. J. Am. Chem. Soc. 133, 15316–15319 (2011).
Li, N., Ramil, C. P., Lim, R. K. V. & Lin, Q. A genetically encoded alkyne directs palladium-mediated protein labeling on live mammalian cell surface. ACS Chem. Biol. 10, 379–384 (2015).
Li, J. et al. Palladium-triggered deprotection chemistry for protein activation in living cells. Nat. Chem. 6, 352–361 (2014).
Wang, J. et al. Chemical remodeling of cell-surface sialic acids through a palladium-triggered bioorthogonal elimination reaction. Angew. Chem. Int. Ed. 54, 5364–5368 (2015).
Yusop, R. M., Unciti-Broceta, A., Johansson, E. M. V., Sánchez-Martin, R. M. & Bradley, M. Palladium-mediated intracellular chemistry. Nat. Chem. 3, 239–243 (2011).
Wang, F., Zhang, Y., Du, Z., Ren, J. & Qu, X. Designed heterogeneous palladium catalysts for reversible light-controlled bioorthogonal catalysis in living cells. Nat. Commun. 9, 1209 (2018).
Tomás-Gamasa, M., Martínez-Calvo, M., Couceiro, J. R. & Mascareñas, J. L. Transition metal catalysis in the mitochondria of living cells. Nat. Commun. 7, 12538 (2016).
Lin, Y. A., Chalker, J. M., Floyd, N., Bernardes, G. J. L. & Davis, B. G. Allyl sulfides are privileged substrates in aqueous cross-metathesis: application to site-selective protein modification. J. Am. Chem. Soc. 130, 9642–9643 (2008).
Lin, Y. A. et al. Rapid cross-metathesis for reversible protein modifications via chemical access to Se-allyl-selenocysteine in proteins. J. Am. Chem. Soc. 135, 12156–12159 (2013).
Bloom, S. et al. Decarboxylative alkylation for site-selective bioconjugation of native proteins via oxidation potentials. Nat. Chem. 10, 205–211 (2018).
Hooker, J. M., Kovacs, E. W. & Francis, M. B. Interior surface modification of bacteriophage MS2. J. Am. Chem. Soc. 126, 3718–3719 (2004).
Sengupta, S. & Chandrasekaran, S. Modifications of amino acids using arenediazonium salts. Org. Biomol. Chem. 17, 8308–8329 (2019).
Addy, P. S., Erickson, S. B., Italia, J. S. & Chatterjee, A. A chemoselective rapid azo-coupling reaction (CRACR) for unclickable bioconjugation. J. Am. Chem. Soc. 139, 11670–11673 (2017).
Lin, S. et al. Redox-based reagents for chemoselective methionine bioconjugation. Science 355, 597–602 (2017).
Taylor, M. T., Nelson, J. E., Suero, M. & Gaunt, M. J. A protein functionalization platform based on selective reactions at methionine residues. Nature 562, 563–568 (2018).
Alvarez-Dorta, D. et al. Electrochemically promoted tyrosine-click-chemistry for protein labeling. J. Am. Chem. Soc. 140, 17120–17126 (2018).
Geng, J. et al. Radical polymerization inside living cells. Nat. Chem. 11, 578–586 (2019).
Vidal, C., Tomás-Gamasa, M., Gutiérrez-González, A. & Mascareñas, J. L. Ruthenium-catalyzed redox isomerizations inside living cells. J. Am. Chem. Soc. 141, 5125–5129 (2019).
Acknowledgements
S.S.N. is an Allergan Graduate Research Fellow. J.A.P. is a Cottrell Scholar, Alfred P. Sloan Fellow and Dreyfus Scholar. This work was funded by the US National Institutes of Health (award no. R01 GM126226). The authors thank members of the Prescher lab for helpful discussions during manuscript preparation.
Author information
Authors and Affiliations
Contributions
The authors contributed equally to all aspects of the article.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information
Nature Reviews Chemistry thanks Q. Lin and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Nguyen, S.S., Prescher, J.A. Developing bioorthogonal probes to span a spectrum of reactivities. Nat Rev Chem 4, 476–489 (2020). https://doi.org/10.1038/s41570-020-0205-0
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41570-020-0205-0
This article is cited by
-
Site-specific bioorthogonal protein labelling by tetrazine ligation using endogenous β-amino acid dienophiles
Nature Chemistry (2023)
-
Reaction profiles for quantum chemistry-computed [3 + 2] cycloaddition reactions
Scientific Data (2023)
-
Recent Advances in Bioorthogonal Ligation and Bioconjugation
Topics in Current Chemistry (2023)
-
Spatiotemporal multiplexed immunofluorescence imaging of living cells and tissues with bioorthogonal cycling of fluorescent probes
Nature Biotechnology (2022)
-
Light-activated tetrazines enable precision live-cell bioorthogonal chemistry
Nature Chemistry (2022)