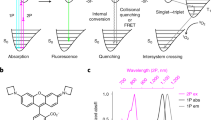
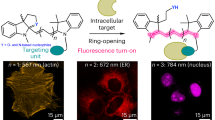
Abstract
Fluorophores have transformed the way we study biological systems, enabling non-invasive studies in cells and intact organisms, which increase our understanding of complex processes at the molecular level. Fluorescent amino acids have become an essential chemical tool because they can be used to construct fluorescent macromolecules, such as peptides and proteins, without disrupting their native biomolecular properties. Fluorescent and fluorogenic amino acids with unique photophysical properties have been designed for tracking protein–protein interactions in situ or imaging nanoscopic events in real time with high spatial resolution. In this Review, we discuss advances in the design and synthesis of fluorescent amino acids and how they have contributed to the field of chemical biology in the past 10 years. Important areas of research that we review include novel methodologies to synthesize building blocks with tunable spectral properties, their integration into peptide and protein scaffolds using site-specific genetic encoding and bioorthogonal approaches, and their application to design novel artificial proteins, as well as to investigate biological processes in cells by means of optical imaging.

This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Chan, J., Dodani, S. C. & Chang, C. J. Reaction-based small-molecule fluorescent probes for chemoselective bioimaging. Nat. Chem. 4, 973–984 (2012).
Lavis, L. D. & Raines, R. T. Bright building blocks for chemical biology. ACS Chem. Biol. 9, 855–866 (2014).
Klymchenko, A. S. Solvatochromic and fluorogenic dyes as environment-sensitive probes: design and biological applications. Acc. Chem. Res. 50, 366–375 (2017).
Park, S. J. et al. Mechanistic elements and critical factors of cellular reprogramming revealed by stepwise global gene expression analyses. Stem Cell Res. 12, 730–741 (2014).
Shimomura, O., Johnson, F. H. & Saiga, Y. Extraction, purification and properties of aequorin, a bioluminescent protein from the luminous hydromedusan, Aequorea. J. Cell. Comp. Physiol. 59, 223–239 (1962).
Tsien, R. Y. The green fluorescent protein. Annu. Rev. Biochem. 67, 509–544 (1998).
Heim, R., Cubitt, A. B. & Tsien, R. Y. Improved green fluorescence. Nature 373, 663–664 (1995).
Keppler, A. et al. A general method for the covalent labeling of fusion proteins with small molecules in vivo. Nat. Biotechnol. 21, 86–89 (2003).
Gautier, A. et al. An engineered protein tag for multiprotein labeling in living cells. Chem. Biol. 15, 128–136 (2008).
Los, G. V. et al. HaloTag: a novel protein labeling technology for cell imaging and protein analysis. ACS Chem. Biol. 3, 373–382 (2008).
Griffin, B. A., Adams, S. R. & Tsien, R. Y. Specific covalent labeling of recombinant protein molecules inside live cells. Science 281, 269–272 (1998).
Martin, B. R., Giepmans, B. N. G., Adams, S. R. & Tsien, R. Y. Mammalian cell–based optimization of the biarsenical-binding tetracysteine motif for improved fluorescence and affinity. Nat. Biotechnol. 23, 1308–1314 (2005).
Krueger, A. T. & Imperiali, B. Fluorescent amino acids: modular building blocks for the assembly of new tools for chemical biology. ChemBioChem 14, 788–799 (2013).
Nitz, M., Mezo, A. R., Ali, M. H. & Imperiali, B. Enantioselective synthesis and application of the highly fluorescent and environment-sensitive amino acid 6-(2-dimethylaminonaphthoyl) alanine (DANA). Chem. Commun. 17, 912–1913 (2002).
Vázquez, M. E., Blanco, J. B. & Imperiali, B. Photophysics and biological applications of the environment-sensitive fluorophore 6-N,N-dimethylamino-2,3-naphthalimide. J. Am. Chem. Soc. 127, 1300–1306 (2005).
Socher, E. & Imperiali, B. FRET-Capture: a sensitive method for the detection of dynamic protein interactions. ChemBioChem 14, 53–57 (2013).
Shults, M. D. & Imperiali, B. Versatile fluorescence probes of protein kinase activity. J. Am. Chem. Soc. 125, 14248–14249 (2003).
Venkatraman, P. et al. Fluorogenic probes for monitoring peptide binding to class II MHC proteins in living cells. Nat. Chem. Biol. 3, 222–228 (2007).
Vázquez, M. E., Rothman, D. M. & Imperiali, B. A new environment-sensitive fluorescent amino acid for Fmoc-based solid phase peptide synthesis. Org. Biomol. Chem. 2, 1965–1966 (2004).
Wang, J., Xie, J. & Schultz, P. G. A genetically encoded fluorescent amino acid. J. Am. Chem. Soc. 128, 8738–8739 (2006).
Kielland, N., Vendrell, M., Lavilla, R. & Chang, Y.-T. Imaging histamine in live basophils and macrophages with a fluorescent mesoionic acid fluoride. Chem. Commun. 48, 7401–7403 (2012).
Weiss, J. T. et al. Extracellular palladium-catalysed dealkylation of 5-fluoro-1-propargyl-uracil as a bioorthogonally activated prodrug approach. Nat. Commun. 5, 3277–3285 (2014).
Ramil, C. P. & Lin, Q. Photoclick chemistry: a fluorogenic light-triggered in vivo ligation reaction. Curr. Opin. Chem. Biol. 21, 89–95 (2014).
Devaraj, N. K. The future of bioorthogonal chemistry. ACS Cent. Sci. 4, 952–959 (2018).
de Moliner, F., Kielland, N., Lavilla, R. & Vendrell, M. Modern synthetic avenues for the preparation of functional fluorophores. Angew. Chem. Int. Ed. 56, 3758–3769 (2017).
Teale, F. W. & Weber, G. Ultraviolet fluorescence of the aromatic amino acids. Biochem. J. 65, 476–482 (1957).
Ghisaidoobe, A. B. T. & Chung, S. J. Intrinsic tryptophan fluorescence in the detection and analysis of proteins: a focus on Förster resonance energy transfer techniques. Int. J. Mol. Sci. 15, 22518–22538 (2014).
Merkel, L., Hoesl, M. G., Albrecht, M., Schmidt, A. & Budisa, N. Blue fluorescent amino acids as in vivo building blocks for proteins. ChemBioChem 11, 305–314 (2010).
Talukder, P. et al. Cyanotryptophans as novel fluorescent probes for studying protein conformational changes and DNA–protein interaction. Biochemistry 54, 7457–7469 (2015).
Hilaire, M. R. et al. Blue fluorescent amino acid for biological spectroscopy and microscopy. Proc. Natl Acad. Sci. USA 114, 6005–6009 (2017). Small analogue of a naturally occurring amino acid that behaves as a non-disruptive building block for the preparation of fluorescent peptides.
Zhang, K. et al. Synthesis and application of the blue fluorescent amino acid l-4-cyanotryptophan to assess peptide–membrane interactions. Chem. Commun. 55, 5095–5098 (2019).
Winn, M., Francis, D. & Micklefield, J. De novo biosynthesis of “non-natural” thaxtomin phytotoxins. Angew. Chem. Int. Ed. 57, 6830–6833 (2018).
Boville, C. E., Romney, D. K., Almhjell, P. J., Sieben, M. & Arnold, F. H. Improved synthesis of 4-cyanotryptophan and other tryptophan analogues in aqueous solvent using variants of TrpB from Thermotoga maritima. J. Org. Chem. 83, 7447–7452 (2018).
Talukder, P., Chen, S., Arce, P. M. & Hecht, S. M. Efficient asymmetric synthesis of tryptophan analogues having useful photophysical properties. Org. Lett. 16, 556–559 (2014).
Wen, J. et al. Highly N2-selective coupling of 1,2,3-triazoles with indole and pyrrole. Chem. Eur. J. 20, 974–978 (2014).
Williams, T. J., Reay, A. J., Whitwood, A. C. & Fairlamb, I. J. S. A mild and selective Pd-mediated methodology for the synthesis of highly fluorescent 2-arylated tryptophans and tryptophan-containing peptides: a catalytic role for Pd0 nanoparticles? Chem. Commun. 50, 3052–3054 (2014).
Bartoccini, F., Bartolucci, S., Mari, M. & Piersanti, G. A simple, modular synthesis of C4-substituted tryptophan derivatives. Org. Biomol. Chem. 14, 10095–10100 (2016).
Talukder, P. et al. Tryptophan-based fluorophores for studying protein conformational changes. Bioorg. Med. Chem. 22, 5924–5934 (2014).
Chen, S. et al. Fluorescent biphenyl derivatives of phenylalanine suitable for protein modification. Biochemistry 52, 8580–8589 (2013).
Chen, S. et al. Detection of dihydrofolate reductase conformational change by FRET using two fluorescent amino acids. J. Am. Chem. Soc. 135, 12924–12927 (2013).
Maity, J., Honcharenko, D. & Strömberg, R. Synthesis of fluorescent d-amino acids with 4-acetamidobiphenyl and 4-N,N-dimethylamino-1,8-naphthalimido containing side chains. Tetrahedron Lett. 56, 4780–4783 (2015).
Cheruku, P. et al. Tyrosine-derived stimuli responsive, fluorescent amino acids. Chem. Sci. 6, 1150–1158 (2015). A toolbox of tyrosine-based FlAAs with tunable emission and reversible pH and redox responses, showing potential for biosensing applications.
Bylińska, I., Guzow, K., Wójcik, J. & Wiczk, W. New non-protienogenic fluorescent amino acids: benzoxazol-5-yl-alanine derivatives containing acetylene unit. Synthesis, spectral and photophysical properties. J. Photochem. Photobiol. A Chem. 364, 679–685 (2018).
Hoppmann, C., Alexiev, U., Irran, E. & Rück-Braun, K. Synthesis and fluorescence of xanthone amino acids. Tetrahedron Lett. 54, 4585–4587 (2013).
Speight, L. C. et al. Efficient synthesis and in vivo incorporation of acridon-2-ylalanine, a fluorescent amino acid for lifetime and Förster resonance energy transfer/luminescence resonance energy transfer studies. J. Am. Chem. Soc. 135, 18806–18814 (2013).
Mendive-Tapia, L. et al. Preparation of a Trp-BODIPY fluorogenic amino acid to label peptides for enhanced live-cell fluorescence imaging. Nat. Protoc. 12, 1588–1619 (2017).
Subiros-Funosas, R. et al. Fluorogenic Trp(redBODIPY) cyclopeptide targeting keratin 1 for imaging of aggressive carcinomas. Chem. Sci. 11, 1368–1374 (2020). Development of an optically enhanced Trp-redBODIPY and validation in cyclic peptides for imaging aggressive carcinomas.
Terrey, M. J., Holmes, A., Perry, C. C. & Cross, W. B. C–H olefination of tryptophan residues in peptides: control of residue selectivity and peptide–amino acid cross-linking. Org. Lett. 21, 7902–7907 (2019).
Wang, W., Lorion, M. M., Martinazzoli, O. & Ackermann, L. BODIPY peptide labeling by late-stage C(sp3)–H activation. Angew. Chem. Int. Ed. 57, 10554–10558 (2018).
Schischko, A. et al. Late-stage peptide C–H alkylation for bioorthogonal C–H activation featuring solid phase peptide synthesis. Nat. Commun. 10, 3553 (2019).
Pereira, G., Vilaça, H. & Ferreira, P. M. T. Synthesis of new β-amidodehydroaminobutyric acid derivatives and of new tyrosine derivatives using copper catalyzed C–N and C–O coupling reactions. Amino Acids 44, 335–344 (2013).
Bag, S. S., Jana, S. & Pradhan, M. K. Synthesis, photophysical properties of triazolyl-donor/acceptor chromophores decorated unnatural amino acids: Incorporation of a pair into Leu-enkephalin peptide and application of triazolylperylene amino acid in sensing BSA. Bioorg. Med. Chem. 24, 3579–3595 (2016).
Benedetti, E., Veliz, A. B. E., Charpenay, M., Kocsis, L. S. & Brummond, K. M. Attachable solvatochromic fluorophores and bioconjugation studies. Org. Lett. 15, 2578–2581 (2013).
Li, C. et al. Click chemistry to fluorescent amino esters: synthesis and spectroscopic studies. Eur. J. Org. Chem. 2010, 2395–2405 (2010).
Ferreira, P. M. T., Monteiro, L. S., Pereira, G., Castanheira, E. M. S. & Frost, C. G. Synthesis of fluorescent alanines by a rhodium-catalysed conjugate addition of arylboronic acids to dehydroalanine derivatives. Eur. J. Org. Chem. 2013, 550–556 (2013).
Hsu, Y.-P. et al. Full color palette of fluorescent d-amino acids for in situ labeling of bacterial cell walls. Chem. Sci. 8, 6313–6321 (2017).
Häußler, D. & Gütschow, M. Fluorescently labeled amino acids as building blocks for bioactive molecules. Synthesis 48, 245–255 (2016).
Kuru, E., Tekkam, S., Hall, E., Brun, Y. V. & Van Nieuwenhze, M. S. Synthesis of fluorescent d-amino acids and their use for probing peptidoglycan synthesis and bacterial growth in situ. Nat. Protoc. 10, 33–52 (2015).
Katritzky, A. R., Ozcan, S. & Todadze, E. Synthesis and fluorescence of the new environment-sensitive fluorophore 6-chloro-2,3-naphthalimide derivative. Org. Biomol. Chem. 8, 1296–1300 (2010).
Esteves, C. I. C., Silva, A. M. F., Raposo, M. M. M. & Costa, S. P. G. Unnatural benz-X-azolyl asparagine derivatives as novel fluorescent amino acids: synthesis and photophysical characterization. Tetrahedron 65, 9373–9377 (2009).
Yokoo, H., Kagechika, H., Ohsaki, A. & Hirano, T. A polarity-sensitive fluorescent amino acid and its incorporation into peptides for the ratiometric detection of biomolecular interactions. ChemPlusChem 84, 1716–1719 (2019).
Shukla, L., Moodie, L. W. K., Kindahl, T. & Hedberg, C. Synthesis and spectroscopic properties of fluorinated coumarin lysine derivatives. J. Org. Chem. 83, 4792–4799 (2018).
Bag, S. S. & De, S. Isothiocyanyl alanine as a synthetic intermediate for the synthesis of thioureayl alanines and subsequent aminotetrazolyl alanines. J. Org. Chem. 82, 12276–12285 (2017).
Mohite, A. R. & Bhat, R. G. Enantiopure synthesis of side chain-modified α-amino acids and 5-cis-alkylprolines. J. Org. Chem. 77, 5423–5428 (2012).
Fowler, L. S., Ellis, D. & Sutherland, A. Synthesis of fluorescent enone derived α-amino acids. Org. Biomol. Chem. 7, 4309–4316 (2009).
Navo, C. D. et al. Cell-penetrating peptides containing fluorescent d-cysteines. Chem. Eur. J. 24, 7991–8000 (2018).
Wörner, S., Rönicke, F., Ulrich, A. S. & Wagenknecht, H.-A. 4-Aminophthalimide amino acids as small and environment-sensitive fluorescent probes for transmembrane peptides. ChemBioChem 21, 618–622 (2020).
Xiang, Z. & Wang, L. Enantiospecific synthesis of genetically encodable fluorescent unnatural amino acid l-3-(6-acetylnaphthalen-2-ylamino)-2-aminopropanoic acid. J. Org. Chem. 76, 6367–6371 (2011).
Zhao, Y., Pirrung, M. C. & Liao, J. A fluorescent amino acid probe to monitor efficiency of peptide conjugation to glass surfaces for high density microarrays. Mol. BioSyst. 8, 879–887 (2012).
Arribat, M., Rémond, E., Clément, S., Lee, A. V. D. & Cavelier, F. Phospholyl(borane) amino acids and peptides: stereoselective synthesis and fluorescent properties with large Stokes shift. J. Am. Chem. Soc. 140, 1028–1034 (2018).
Strizhak, A. V. et al. Two-color fluorescent l-amino acid mimic of tryptophan for probing peptide–nucleic acid complexes. Bioconjugate Chem. 23, 2434–2443 (2012). First report of a tyrosine-based FlAA exhibiting excited-state intramolecular proton transfer and hydration-sensitive dual emission.
Postupalenko, V. Y. et al. Dual-fluorescence l-amino acid reports insertion and orientation of melittin peptide in cell membranes. Bioconjugate Chem. 24, 1998–2007 (2013).
Sholokh, M. et al. Fluorescent amino acid undergoing excited state intramolecular proton transfer for site-specific probing and imaging of peptide interactions. J. Phys. Chem. B 119, 2585–2595 (2015).
Taki, M., Yamazaki, Y., Suzuki, Y. & Sisido, M. Introduction of a highly photodurable and common-laser excitable fluorescent amino acid into a peptide as a FRET acceptor for protease cleavage detection. Chem. Lett. 39, 818–819 (2010).
Bell, J. D. et al. Synthesis and photophysical properties of benzotriazole-derived unnatural α-amino acids. J. Org. Chem. 84, 10436–10448 (2019).
Gilfillan, L., Artschwager, R., Harkiss, A. H., Liskamp, R. M. J. & Sutherland, A. Synthesis of pyrazole containing α-amino acids via a highly regioselective condensation/aza-Michael reaction of β-aryl α,β-unsaturated ketones. Org. Biomol. Chem. 13, 4514–4523 (2015).
Harkiss, A. H., Bell, J. D., Knuhtsen, A., Jamieson, A. G. & Sutherland, A. Synthesis and fluorescent properties of β-pyridyl α-amino acids. J. Org. Chem. 84, 2879–2890 (2019).
Bell, J. D. et al. Conformationally rigid pyrazoloquinazoline α-amino acids: one- and two-photon induced fluorescence. Chem. Commun. 56, 1887–1890 (2020).
Häußler, D. & Gütschow, M. Synthesis of a fluorescent-labeled bisbenzamidine containing the central (6,7-dimethoxy-4-coumaryl)alanine building block. Heteroat. Chem. 26, 367–373 (2015).
Koopmans, T., van Haren, M., van Ufford, L. Q., Beekman, J. M. & Martin, N. I. A concise preparation of the fluorescent amino acid l-(7-hydroxycoumarin-4-yl) ethylglycine and extension of its utility in solid phase peptide synthesis. Bioorg. Med. Chem. 21, 553–559 (2013).
Moodie, L. W. K., Chammaa, S., Kindahl, T. & Hedberg, C. Palladium-mediated approach to coumarin-functionalized amino acids. Org. Lett. 19, 2797–2800 (2017).
Fernandez, A., Thompson, E. J., Pollard, J. W., Kitamura, T. & Vendrell, M. A fluorescent activatable AND-gate chemokine CCL2 enables in vivo detection of metastasis-associated macrophages. Angew. Chem. Int. Ed. 58, 16894–16898 (2019).
Joshi, P. N. & Rai, V. Single-site labeling of histidine in proteins, on-demand reversibility, and traceless metal-free protein purification. Chem. Commun. 55, 1100–1103 (2019).
Cheng, M. H. Y., Savoie, H., Bryden, F. & Boyle, R. W. A convenient method for multicolour labelling of proteins with BODIPY fluorophores via tyrosine residues. Photochem. Photobiol. Sci. 16, 1260–1267 (2017).
Zhao, C. et al. Searching for the optimal fluorophore to label antimicrobial peptides. ACS Comb. Sci. 18, 689–696 (2016).
Vendrell, M. et al. Biotin ergopeptide probes for dopamine receptors. J. Med. Chem. 54, 1080–1090 (2011).
Palomo, J. M. Solid-phase peptide synthesis: an overview focused on the preparation of biologically relevant peptides. RSC Adv. 4, 32658–32672 (2014).
Sainlos, M., Iskenderian, W. S. & Imperiali, B. A general screening strategy for peptide-based fluorogenic ligands: probes for dynamic studies of PDZ domain-mediated interactions. J. Am. Chem. Soc. 131, 6680–6682 (2009). Pioneering work on solvatochromic phthalimide amino acids and their integration into peptide structures to study dynamic protein–protein interactions.
Loving, G. & Imperiali, B. A versatile amino acid analogue of the solvatochromic fluorophore 4-N,N-dimethylamino-1,8-naphthalimide: a powerful tool for the study of dynamic protein interactions. J. Am. Chem. Soc. 130, 13630–13638 (2008).
Wang, W. et al. A fluorogenic peptide probe developed by in vitro selection using tRNA carrying a fluorogenic amino acid. Chem. Commun. 50, 2962–2964 (2014).
Wang, W. et al. Fluorogenic enhancement of an in vitro-selected peptide ligand by replacement of a fluorescent group. Anal. Chem. 88, 7991–7997 (2016).
Heru, C., Jurgen, S., Tino, Z. & Horst, A. Fluorescent analogues of the insect neuropeptide helicokinin I: synthesis, photophysical characterization and biological activity. Prot. Pept. Lett. 17, 431–436 (2010).
Manandhar, Y. et al. In vitro selection of a peptide aptamer that changes fluorescence in response to verotoxin. Biotechnol. Lett. 37, 619–625 (2015).
Newton, L. D., Pascu, S. I., Tyrrell, R. M. & Eggleston, I. M. Development of a peptide-based fluorescent probe for biological heme monitoring. Org. Biomol. Chem. 17, 467–471 (2019).
Zhao, C., Mendive-Tapia, L. & Vendrell, M. Fluorescent peptides for imaging of fungal cells. Arch. Biochem. Biophys. 661, 187–195 (2019).
Mendive-Tapia, L. et al. Spacer-free BODIPY fluorogens in antimicrobial peptides for direct imaging of fungal infection in human tissue. Nat. Commun. 7, 10940 (2016). First report of Trp-BODIPY as a fluorogenic amino acid to non-invasively label peptides for live-cell and ex vivo tissue imaging.
Akram, A. R. et al. Enhanced avidity from a multivalent fluorescent antimicrobial peptide enables pathogen detection in a human lung model. Sci. Rep. 9, 8422 (2019).
Subiros-Funosas, R. et al. A Trp-BODIPY cyclic peptide for fluorescence labelling of apoptotic bodies. Chem. Commun. 53, 945–948 (2017).
Ge, J., Li, L. & Yao, S. Q. A self-immobilizing and fluorogenic unnatural amino acid that mimics phosphotyrosine. Chem. Commun. 47, 10939–10941 (2011).
TB, K. C. et al. Wash-free and selective imaging of epithelial cell adhesion molecule (EpCAM) expressing cells with fluorogenic peptide ligands. Biochem. Biophys. Res. Commun. 500, 283–287 (2018).
Maeno, T. et al. Targeted delivery of fluorogenic peptide aptamers into live microalgae by femtosecond laser photoporation at single-cell resolution. Sci. Rep. 8, 8271 (2018).
Egan, A. J. F., Cleverley, R. M., Peters, K., Lewis, R. J. & Vollmer, W. Regulation of bacterial cell wall growth. FEBS J. 284, 851–867 (2017).
Dörr, T., Moynihan, P. J. & Mayer, C. Bacterial cell wall structure and dynamics. Front. Microbiol. 10, 2051 (2019).
Radkov, A. D., Hsu, Y.-P., Booher, G. & VanNieuwenhze, M. S. Imaging bacterial cell wall biosynthesis. Annu. Rev. Biochem. 87, 991–1014 (2018).
Hsu, Y.-P., Booher, G., Egan, A., Vollmer, W. & VanNieuwenhze, M. S. d-Amino acid derivatives as in situ probes for visualizing bacterial peptidoglycan biosynthesis. Acc. Chem. Res. 52, 2713–2722 (2019).
de Pedro, M. A., Quintela, J. C., Höltje, J. V. & Schwarz, H. Murein segregation in Escherichia coli. J. Bacteriol. 179, 2823–2834 (1997).
Cava, F., de Pedro, M. A., Lam, H., Davis, B. M. & Waldor, M. K. Distinct pathways for modification of the bacterial cell wall by non-canonical d-amino acids. EMBO J. 30, 3442–3453 (2011).
Lupoli, T. J. et al. Transpeptidase-mediated incorporation of d-amino acids into bacterial peptidoglycan. J. Am. Chem. Soc. 133, 10748–10751 (2011).
Siegrist, M. S. et al. d-Amino acid chemical reporters reveal peptidoglycan dynamics of an intracellular pathogen. ACS Chem. Biol. 8, 500–505 (2013).
Kuru, E. et al. In situ probing of newly synthesized peptidoglycan in live bacteria with fluorescent d-amino acids. Angew. Chem. Int. Ed. 51, 12519–12523 (2012). Development of a toolbox of fluorescent d-amino acids for labelling peptidoglycans and monitoring bacterial cell-wall growth.
Liechti, G. W. et al. A new metabolic cell-wall labelling method reveals peptidoglycan in Chlamydia trachomatis. Nature 506, 507–510 (2014).
Hudak, J. E., Alvarez, D., Skelly, A., von Andrian, U. H. & Kasper, D. L. Illuminating vital surface molecules of symbionts in health and disease. Nat. Microbiol. 2, 17099 (2017).
Wang, W. et al. Assessing the viability of transplanted gut microbiota by sequential tagging with d-amino acid-based metabolic probes. Nat. Commun. 10, 1317 (2019).
Hu, F. et al. Visualization and in situ ablation of intracellular bacterial pathogens through metabolic labeling. Angew. Chem. Int. Ed. https://doi.org/10.1002/anie.201910187 (2019).
Kuru, E. Mechanisms of incorporation for d-amino acid probes that target peptidoglycan biosynthesis. ACS Chem. Biol. 14, 2745–2756 (2019).
Kuru, E. et al. Fluorescent d-amino-acids reveal bi-cellular cell wall modifications important for Bdellovibrio bacteriovorus predation. Nat. Microbiol. 2, 1648–1657 (2017).
Morales Angeles, D. et al. Pentapeptide-rich peptidoglycan at the Bacillus subtilis cell-division site. Mol. Microbiol. 104, 319–333 (2017).
Baranowski, C. et al. Maturing Mycobacterium smegmatis peptidoglycan requires non-canonical crosslinks to maintain shape. eLife 7, e37516 (2018).
Bisson-Filho, A. W. et al. Treadmilling by FtsZ filaments drives peptidoglycan synthesis and bacterial cell division. Science 355, 739–743 (2017).
Yang, X. et al. GTPase activity–coupled treadmilling of the bacterial tubulin FtsZ organizes septal cell wall synthesis. Science 355, 744–747 (2017).
Liechti, G. et al. Pathogenic Chlamydia lack a classical sacculus but synthesize a narrow, mid-cell peptidoglycan ring, regulated by MreB, for cell division. PLoS Pathog. 12, e1005590 (2016).
Hsu, Y.-P. et al. Fluorogenic d-amino acids enable real-time monitoring of peptidoglycan biosynthesis and high-throughput transpeptidation assays. Nat. Chem. 11, 335–341 (2019). Rotor-fluorogenic d-amino acids for real-time visualization of transpeptidase reactions and high-throughput screening of antibacterial drugs.
Rodriguez, E. A. et al. The growing and glowing toolbox of fluorescent and photoactive proteins. Trends Biochem. Sci. 42, 111–129 (2017).
Jing, C. & Cornish, V. W. Chemical tags for labeling proteins inside living cells. Acc. Chem. Res. 44, 784–792 (2011).
Spicer, C. D. & Davis, B. G. Selective chemical protein modification. Nat. Commun. 5, 4740 (2014).
Kajihara, D. et al. FRET analysis of protein conformational change through position-specific incorporation of fluorescent amino acids. Nat. Methods 3, 923–929 (2006).
Goto, Y., Katoh, T. & Suga, H. Flexizymes for genetic code reprogramming. Nat. Protoc. 6, 779–790 (2011).
Lang, K. & Chin, J. W. Cellular incorporation of unnatural amino acids and bioorthogonal labeling of proteins. Chem. Rev. 114, 4764–4806 (2014).
Young, D. D. & Schultz, P. G. Playing with the molecules of life. ACS Chem. Biol. 13, 854–870 (2018).
Chin, J. W. Expanding and reprogramming the genetic code of cells and animals. Annu. Rev. Biochem. 83, 379–408 (2014).
Chin, J. W. Expanding and reprogramming the genetic code. Nature 550, 53–60 (2017).
Dumas, A., Lercher, L., Spicer, C. D. & Davis, B. G. Designing logical codon reassignment – expanding the chemistry in biology. Chem. Sci. 6, 50–69 (2015).
Fredens, J. et al. Total synthesis of Escherichia coli with a recoded genome. Nature 569, 514–518 (2019).
Wang, K. et al. Optimized orthogonal translation of unnatural amino acids enables spontaneous protein double-labelling and FRET. Nat. Chem. 6, 393–403 (2014).
Sachdeva, A., Wang, K., Elliott, T. & Chin, J. W. Concerted, rapid, quantitative, and site-specific dual labeling of proteins. J. Am. Chem. Soc. 136, 7785–7788 (2014).
Lampkowski, J. S., Uthappa, D. M. & Young, D. D. Site-specific incorporation of a fluorescent terphenyl unnatural amino acid. Bioorg. Med. Chem. Lett. 25, 5277–5280 (2015).
Kuhn, S. M., Rubini, M., Müller, M. A. & Skerra, A. Biosynthesis of a fluorescent protein with extreme pseudo-Stokes shift by introducing a genetically encoded non-natural amino acid outside the fluorophore. J. Am. Chem. Soc. 133, 3708–3711 (2011).
Lacey, V. K. et al. A fluorescent reporter of the phosphorylation status of the substrate protein STAT3. Angew. Chem. Int. Ed. 50, 8692–8696 (2011).
Amaro, M. et al. Site-specific analysis of protein hydration based on unnatural amino acid fluorescence. J. Am. Chem. Soc. 137, 4988–4992 (2015).
Steinberg, X. et al. Conformational dynamics in TRPV1 channels reported by an encoded coumarin amino acid. eLife 6, e28626 (2017).
Luo, J. et al. Genetically encoded optochemical probes for simultaneous fluorescence reporting and light activation of protein function with two-photon excitation. J. Am. Chem. Soc. 136, 15551–15558 (2014).
Wan, W., Tharp, J. M. & Liu, W. R. Pyrrolysyl-tRNA synthetase: an ordinary enzyme but an outstanding genetic code expansion tool. Biochim. Biophys. Acta 1844, 1059–1070 (2014).
Srinivasan, G., James, C. M. & Krzycki, J. A. Pyrrolysine encoded by UAG in archaea: charging of a UAG-decoding specialized tRNA. Science 296, 1459–1462 (2002).
Elliott, T. S. et al. Proteome labeling and protein identification in specific tissues and at specific developmental stages in an animal. Nat. Biotechnol. 32, 465–472 (2014).
Summerer, D. et al. A genetically encoded fluorescent amino acid. Proc. Natl Acad. Sci. USA 103, 9785–9789 (2006).
Lee, H. S., Guo, J., Lemke, E. A., Dimla, R. D. & Schultz, P. G. Genetic incorporation of a small, environmentally sensitive, fluorescent probe into proteins in Saccharomyces cerevisiae. J. Am. Chem. Soc. 131, 12921–12923 (2009). Seminal work on the genetic encoding of the environmentally sensitive amino acid ANAP into protein structures.
Chatterjee, A., Guo, J., Lee, H. S. & Schultz, P. G. A genetically encoded fluorescent probe in mammalian cells. J. Am. Chem. Soc. 135, 12540–12543 (2013).
Chen, R. F. Fluorescence of dansyl amino acids in organic solvents and protein solutions. Arch. Biochem. Biophys. 120, 609–620 (1967).
Kalstrup, T. & Blunck, R. Dynamics of internal pore opening in Kv channels probed by a fluorescent unnatural amino acid. Proc. Natl Acad. Sci. USA 110, 8272–8277 (2013). The incorporation of ANAP into proteins enabled the investigation of the gating mechanism of voltage-gated potassium channels.
Sakata, S., Jinno, Y., Kawanabe, A. & Okamura, Y. Voltage-dependent motion of the catalytic region of voltage-sensing phosphatase monitored by a fluorescent amino acid. Proc. Natl Acad. Sci. USA 113, 7521–7526 (2016).
Park, S.-H., Ko, W., Lee, H. S. & Shin, I. Analysis of protein–protein interaction in a single live cell by using a FRET system based on genetic code expansion technology. J. Am. Chem. Soc. 141, 4273–4281 (2019).
Preciado, S. et al. Synthesis and biological evaluation of a post-synthetically modified Trp-based diketopiperazine. MedChemComm 4, 1171–1174 (2013).
Mendive-Tapia, L. et al. New peptide architectures through C–H activation stapling between tryptophan–phenylalanine/tyrosine residues. Nat. Commun. 6, 7160 (2015).
Chen, X. et al. Histidine-specific peptide modification via visible-light-promoted C–H alkylation. J. Am. Chem. Soc. 141, 18230–18237 (2019).
Peciak, K., Laurine, E., Tommasi, R., Choi, J.-W. & Brocchini, S. Site-selective protein conjugation at histidine. Chem. Sci. 10, 427–439 (2019).
Ban, H., Gavrilyuk, J. & Barbas, C. F. Tyrosine bioconjugation through aqueous ene-type reactions: a click-like reaction for tyrosine. J. Am. Chem. Soc. 132, 1523–1525 (2010).
Tilley, S. D. & Francis, M. B. Tyrosine-selective protein alkylation using π-allylpalladium complexes. J. Am. Chem. Soc. 128, 1080–1081 (2006).
Benson, S. et al. SCOTfluors: small, conjugatable, orthogonal, and tunable fluorophores for in vivo imaging of cell metabolism. Angew. Chem. Int. Ed. 58, 6911–6915 (2019).
Su, L. et al. Cu(I)-catalyzed 6-endo-dig cyclization of terminal alkynes, 2-bromoaryl ketones, and amides toward 1-naphthylamines: applications and photophysical properties. J. Am. Chem. Soc. 141, 2535–2544 (2019).
Mellanby, R. J. et al. Tricarbocyanine N-triazoles: the scaffold-of-choice for long-term near-infrared imaging of immune cells in vivo. Chem. Sci. 9, 7261–7270 (2018).
Cosco, E. D. et al. Flavylium polymethine fluorophores for near- and shortwave infrared imaging. Angew. Chem. Int. Ed. 56, 13126–13129 (2017).
Tang, J. et al. Single-atom fluorescence switch: a general approach toward visible-light-activated dyes for biological imaging. J. Am. Chem. Soc. 141, 14699–14706 (2019).
Zheng, Q. et al. Rational design of fluorogenic and spontaneously blinking labels for super-resolution imaging. ACS Cent. Sci. 5, 1602–1613 (2019).
Raymo, F. M. Photoactivatable synthetic fluorophores. Phys. Chem. Chem. Phys. 15, 14840–14850 (2013).
Zhang, Y., Tang, S., Sansalone, L., Baker, J. D. & Raymo, F. M. A photoswitchable fluorophore for the real-time monitoring of dynamic events in living organisms. Chem. Eur. J. 22, 15027–15034 (2016).
Hendricks, J. A. et al. Synthesis of [18F]BODIPY: bifunctional reporter for hybrid optical/positron emission tomography imaging. Angew. Chem. Int. Ed. 51, 4603–4606 (2012).
Zhou, E. Y., Knox, H. J., Liu, C., Zhao, W. & Chan, J. A conformationally restricted aza-BODIPY platform for stimulus-responsive probes with enhanced photoacoustic properties. J. Am. Chem. Soc. 141, 17601–17609 (2019).
Onogi, S. et al. In situ real-time imaging of self-sorted supramolecular nanofibres. Nat. Chem. 8, 743–752 (2016).
Beesley, J. L. & Woolfson, D. N. The de novo design of α-helical peptides for supramolecular self-assembly. Curr. Opin. Biotechnol. 58, 175–182 (2019).
Davis, L. & Greiss, S. Genetic encoding of unnatural amino acids in C. elegans. Methods Mol. Biol. 1728, 389–408 (2018).
Bridge, T. et al. Site-specific encoding of photoactivity in antibodies enables light-mediated antibody–antigen binding on live cells. Angew. Chem. Int. Ed. 58, 17986–17993 (2019).
Ren, T.-B. et al. A general method to increase Stokes shift by introducing alternating vibronic structures. J. Am. Chem. Soc. 140, 7716–7722 (2018).
Acknowledgements
E.K. acknowledges funding of a fellowship from the Life Sciences Research Foundation. A.S. acknowledges funding from the Wellcome Trust (204593/Z/16/Z) and the Biotechnology and Biological Sciences Research Council (BB/R004692/1). M.V. acknowledges funding from an ERC Consolidator Grant (771443). The authors thank S. Shaikh for the useful comments and technical support with the graphical illustrations.
Author information
Authors and Affiliations
Contributions
All authors contributed equally to the preparation of this manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information
Nature Reviews Chemistry thanks A. Klymchenko and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Cheng, Z., Kuru, E., Sachdeva, A. et al. Fluorescent amino acids as versatile building blocks for chemical biology. Nat Rev Chem 4, 275–290 (2020). https://doi.org/10.1038/s41570-020-0186-z
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41570-020-0186-z
This article is cited by
-
Tracking endogenous proteins based on RNA editing-mediated genetic code expansion
Nature Chemical Biology (2024)
-
Modular and diverse synthesis of amino acids via asymmetric decarboxylative protonation of aminomalonic acids
Nature Chemistry (2023)
-
Non-thermal Approach for Electromagnetic Field Exposure to Unfold Heat-Resistant Sunflower Protein
Food and Bioprocess Technology (2023)
-
Development of Orthogonal Aminoacyl tRNA Synthetase Mutant with Enhanced Incorporation Ability with Para-azido-L-phenylalanine
Biotechnology and Bioprocess Engineering (2023)
-
Determination and Imaging of Small Biomolecules and Ions Using Ruthenium(II) Complex-Based Chemosensors
Topics in Current Chemistry (2022)