Abstract
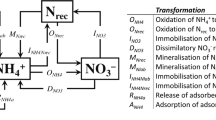
The nitrogen cycle is one of the most important biogeochemical cycles on Earth because nitrogen is an essential nutrient for all life forms. To supplement natural nitrogen fixation, farmers add large amounts of nitrogen-containing fertilizer to their soils such that nitrogen never becomes a limiting nutrient for plant growth. However, of the nitrogen added to fields — most of which is in the form of NH3 and NO3− — only 30–50% is taken up by plants, while the remainder is metabolized by soil microorganisms in processes with detrimental environmental impacts. The first of these processes, that is, nitrification, refers to the biological oxidation of NH3 to NO2− and NO3−, which have low retention in soil and pollute waterways, leading to downstream eutrophication and ultimately ‘dead zones’ (low oxygen zones) in coastal waters, for example, the Gulf of Mexico. In a second process, namely, denitrification, NO3− and NO2− undergo stepwise reduction to N2O and N2. Substantial amounts of the N2O produced in this process escape into the atmosphere, contributing to climate change and ozone destruction. Recent results suggest that nitrification also affords N2O. This Review describes the enzymes involved in NH3 oxidation and N2O production and degradation in the nitrogen cycle. We pay particular attention to the active site structures, the associated coordination chemistry that enables the chemical transformations and the reaction mechanisms.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout








Similar content being viewed by others
References
Lehnert, N., Coruzzi, G., Hegg, E., Seefeldt, L. & Stein, L. NSF Workshop Report: Feeding the World in the 21st Century: Grand Challenges in the Nitrogen Cycle (National Science Foundation: Arlington, VA, 2016).
Canfield, D. E., Glazer, A. N. & Falkowski, P. G. The evolution and future of earth’s nitrogen cycle. Science 330, 192–196 (2010).
Galloway, J. N. et al. Transformation of the nitrogen cycle: recent trends, questions, and potential solutions. Science 320, 889–892 (2008).
Fields, S. Global nitrogen: cycling out of control. Environ. Health Perspect. 112, A556–A563 (2004).
Smil, V. Nitrogen cycle and world food production. World Agric. 2, 9–13 (2011).
Smil, V. Enriching the Earth: Fritz Haber, Carl Bosch and the Transformation of World Food Production (MIT Press, Cambridge, MA, 2001).
Burgess, B. K. & Lowe, D. J. Mechanism of molybdenum nitrogenase. Chem. Rev. 96, 2983–3012 (1996).
Hoffman, B. M., Lukoyanov, D., Yang, Y.-Z., Dean, D. R. & Seefeldt, L. C. Mechanism of nitrogen fixation by nitrogenase: the next stage. Chem. Rev. 114, 4041–4062 (2014).
Burford, R. J. & Fryzuk, M. D. Examining the relationship between coordination mode and reactivity of dinitrogen. Nat. Rev. Chem. 1, 0026 (2017).
Erisman, J. W., Sutton, M. A., Galloway, J., Klimont, Z. & Winiwarter, W. How a century of ammonia synthesis changed the world. Nat. Geosci. 1, 636–639 (2008).
Godfray, H. C. J. et al. Food security: the challenge of feeding 9 billion people. Science 327, 812–818 (2010).
Ferguson, S. J. Nitrogen cycle enzymology. Curr. Opin. Chem. Biol. 2, 182–193 (1998).
Richardson, D. J. & Watmough, N. J. Inorganic nitrogen metabolism in bacteria. Curr. Opin. Chem. Biol. 3, 207–219 (1999).
Maia, L. B. & Moura, J. J. G. How biology handles nitrite. Chem. Rev. 114, 5273–5357 (2014).
Kuypers, M. M. M., Marchant, H. K. & Kartal, B. The microbial nitrogen-cycling network. Nat. Rev. Microbiol. 16, 263–276 (2018).
Ward, B. B. in Nitrification (eds Ward, B. B., Arp, D. J. & Klotz, M.) 3–8 (ASM Press, Sterling, VA, 2011).
Stein, L. Y. & Klotz, M. G. Primer: the nitrogen cycle. Curr. Biol. 26, R94–R98 (2016).
Galloway, J. N., Leach, A. M., Bleeker, A. & Erisman, J. W. A chronology of human understanding of the nitrogen cycle. Phil. Trans. R. Soc. B 368, 20130120 (2013).
Bourzac, K. Cleaning up nitrogen on farm lands. Chem. Eng. News 95, 16–19 (2017).
Hooper, A. B., Arciero, D., Bergmann, D. & Hendrich, M. P. in Respiration in Archaea and Bacteria (ed. Zannoni, D.) 121–147 (Springer, Dordrecht, 2005).
Hooper, A. B. & Terry, K. R. Hydroxylamine oxidoreductase of nitrosomonas: production of nitric oxide from hydroxylamine. Biochim. Biophys. Acta. 571, 12–20 (1979).
van de Graaf, A. A. et al. Anaerobic oxidation of ammonium is a biologically mediated process. Appl. Environ. Microbiol. 61, 1246–1251 (1995).
Kartal, B., Geerts, W. & Jetten, M. S. M. Cultivation, detection, and ecophysiology of anaerobic ammonium-oxidizing bacteria. Methods Enzymol. 486, 89–108 (2011).
Kartal, B. et al. Molecular mechanism of anaerobic ammonium oxidation. Nature 479, 127–130 (2011).
Kartal, B. & Keltjens, J. T. Anammox biochemistry: a tale of heme c proteins. Trends Biochem. Sci. 41, 998–1011 (2016).
van de Vossenberg, J. et al. The metagenome of the marine anammox bacterium ‘Candidatus Scalindua profunda’ illustrates the versatility of this globally important nitrogen cycle bacterium. Environ. Microbiol. 15, 1275–1289 (2013).
Daisuke, H. et al. Anammox organism KSU-1 expresses a NirK-type copper-containing nitrite reductase instead of a NirS-type with cytochrome cd 1. FEBS Lett. 586, 1658–1663 (2012).
Dietl, A. et al. The inner workings of the hydrazine synthase multiprotein complex. Nature 527, 394–397 (2015).
Maalcke, W. J. et al. Characterization of anammox hydrazine dehydrogenase, a key N2-producing enzyme in the global nitrogen cycle. J. Biol. Chem. https://doi.org/10.1074/jbc.M116.735530 (2016).
Lam, P. & Kuypers, M. M. M. Microbial nitrogen cycling processes in oxygen minimum Zones. Annu. Rev. Mar. Sci. 3, 317–345 (2011).
Hu, Z. et al. Nitrogen removal by a nitritation-anammox bioreactor at low temperature. Appl. Env. Microbiol. 79, 2807–2812 (2013).
Zumft, W. G. Cell biology and molecular basis of denitrification. Microbiol. Mol. Biol. Rev. 61, 533–616 (1997).
Moura, I. & Moura, J. J. G. Structural aspects of denitrifying enzymes. Curr. Opin. Chem. Biol. 5, 168–175 (2001).
US Environmental Protection Agency. Overview of greenhouse gases. EPA.gov https://www.epa.gov/ghgemissions/overview-greenhouse-gases (2016).
Kandeler, E., Poll, C., Frankenberger Jr, W. T. & Tabatabai, M. A. in Methods in Soil Enzymology (ed. Dick, R. P.) (Soil Science Society of America, Madison, 2011).
Hodge, A., Robinson, D. & Fitter, A. Are microorganisms more effective than plants at competing for nitrogen? Trends Plant Sci. 5, 304–308 (2000).
Masclaux-Daubresse, C. et al. Nitrogen uptake, assimilation and remobilization in plants: challenges for sustainable and productive agriculture. Ann. Bot. 105, 1141–1157 (2010).
Chain, P. et al. Complete genome sequence of the ammonia-oxidizing bacterium and obligate chemolithoautotroph Nitrosomonas europaea. J. Bacteriol. 185, 2759–2773 (2003).
Heil, J., Vereecken, H. & Brüggemann, N. A review of chemical reactions of nitrification intermediates and their role in nitrogen cycling and nitrogen trace gas formation in soil. Eur. J. Soil Sci. 67, 23–39 (2016).
Gilch, S., Meyer, O. & Schmidt, I. A soluble form of ammonia monooxygenase in Nitrosomonas europaea. Biol. Chem. 390, 863–873 (2009).
Robertson, G. P. & Groffmann, P. M. in Soil Microbiology, Ecology, and Biochemistry (ed. Eldor, P.) 341–364 (Elsevier, Amsterdam, 2007).
Zahn, J. A. & DiSpirito, A. A. Membrane-associated methane monooxygenase from Methylococcus capsulatus (Bath). J. Bacteriol. 178, 1018–1029 (1996).
Hooper, A. B. & Terry, K. R. Specific inhibitors of ammonia oxidation in Nitrosomonas. J. Bacteriol. 115, 480–485 (1973).
Ensign, S. A., Hyman, M. R. & Arp, D. J. In vitro activation of ammonia monooxygenase from Nitrosomonas europaea by copper. J. Bacteriol. 175, 1971–1980 (1993).
Zahn, J. A., Arciero, D. M., Hooper, A. B. & DiSpirito, A. A. Evidence for an iron center in the ammonia monooxygenase from Nitrosomonas europaea. FEBS Lett. 397, 35–38 (1996).
Hooper, A. B., Terry, K. R. & Maxwell, P. C. Hydroxylamine oxidoreductase of Nitrosomonas. Oxidation of diethyldithiocarbamate concomitant with stimulation of nitrite synthesis. Biochim. Biophys. Acta, Bioenerg. 462, 141–152 (1977).
Ramaiah, A. & Nicholas, D. J. D. The synthesis of ATP and the incorporation of 32P by cell-free preparations from Nitrosomonas europaea. Biochim. Biophys. Acta, Gen. Subj. 86, 459–465 (1964).
Igarashi, N., Moriyama, H., Fujiwara, T., Fukumori, Y. & Tanaka, N. The 2.8 Å structure of hydroxylamine oxidoreductase from a nitrifying chemoautotrophic bacterium. Nitrosomonas europaea. Nat. Struct. Biol. 4, 276 (1997).
Cedervall, P., Hooper, A. B. & Wilmot, C. M. Structural studies of hydroxylamine oxidoreductase reveal a unique heme cofactor and a previously unidentified interaction partner. Biochemistry 52, 6211–6218 (2013).
Collins, M. J., Arciero, D. M. & Hooper, A. B. Optical spectropotentiometric resolution of the hemes of hydroxylamine oxidoreductase. Heme quantitation and pH dependence of E m*. J. Biol. Chem. 268, 14655–14662 (1993).
Arciero, D. M., Golombek, A., Hendrich, M. P. & Hooper, A. B. Correlation of optical and EPR signals with the P460 heme of hydroxylamine oxidoreductase from Nitrosomonas europaea. Biochemistry 37, 523–529 (1998).
Caranto, J. D. & Lancaster, K. M. Nitric oxide is an obligate bacterial nitrification intermediate produced by hydroxylamine oxidoreductase. Proc. Natl Acad. Sci. USA 114, 8217–8222 (2017).
Cabail, M. Z. & Pacheco, A. A. Selective one-electron reduction of Nitrosomonas europaea hydroxylamine oxidoreductase with nitric oxide. Inorg. Chem. 42, 270–272 (2003).
Fernández, M. L., Estrin, D. A. & Bari, S. E. Theoretical insight into the hydroxylamine oxidoreductase mechanism. J. Inorg. Biochem. 102, 1523–1530 (2008).
Arciero, D. M., Balny, C. & Hooper, A. B. Spectroscopic and rapid kinetic studies of reduction of cytochrome c554 by hydroxylamine oxidoreductase from Nitrosomonas europaea. Biochemistry 30, 11466–11472 (1991).
Walker, F. A. Nitric oxide interaction with insect nitrophorins and thoughts on the electron configuration of the {FeNO}6 complex. J. Inorg. Biochem. 99, 216–236 (2005).
Maalcke, W. J. et al. Structural basis of biological NO generation by octaheme oxidoreductases. J. Biol. Chem. 289, 1228–1242 (2014).
Pearson, A. R. et al. The crystal structure of cytochrome P460 of Nitrosomonas europaea reveals a novel cytochrome fold and heme−protein cross-link. Biochemistry 46, 8340–8349 (2007).
Andersson, K. K., Kent, T. A., Lipscomb, J. D., Hooper, A. B. & Münck, E. Mössbauer, EPR, and optical studies of the P-460 center of hydroxylamine oxidoreductase from Nitrosomonas. A ferrous heme with an unusually large quadrupole splitting. J. Biol. Chem. 259, 6833–6840 (1984).
Erickson, R. H. & Hooper, A. B. Preliminary characterization of a variant co-binding heme protein from Nitrosomonas. Biochim. Biophys. Acta, Bioenerg. 275, 231–244 (1972).
Miller, D. J., Wood, P. M. & Nicholas, D. J. D. Further characterization of cytochrome P-460 in Nitrosomonas europaea. Microbiology 130, 3049–3054 (1984).
Caranto, J. D., Vilbert, A. C. & Lancaster, K. M. Nitrosomonas europaea cytochrome P460 is a direct link between nitrification and nitrous oxide emission. Proc. Natl Acad. Sci. USA 113, 14704–14709 (2016).
Vilbert, A. C., Caranto, J. D. & Lancaster, K. Influences of the heme–lysine crosslink in cytochrome P460 over redox catalysis and nitric oxide sensitivity. Chem. Sci. 9, 368–379 (2018).
Goodrich, L. E. et al. Electronic structure and biologically relevant reactivity of low-spin {FeNO}8 porphyrin model complexes: new insight from a bis-picket fence porphyrin. Inorg. Chem. 52, 7766–7780 (2013).
Berto, T. C., Praneeth, V. K. K., Goodrich, L. E. & Lehnert, N. Iron–porphyrin NO complexes with covalently attached N-donor ligands: formation of a stable six-coordinate species in solution. J. Am. Chem. Soc. 131, 17116–17126 (2009).
Lehnert, N. et al. Oriented single-crystal nuclear resonance vibrational spectroscopy of [Fe(TPP)(MI)(NO)]: quantitative assessment of the trans effect of NO. Inorg. Chem. 49, 7197–7215 (2010).
Hunt, A. P. & Lehnert, N. Heme–nitrosyls: electronic structure implications for function in biology. Acc. Chem. Res. 48, 2117–2125 (2015).
Herzik Jr., M. A., Jonnalagadda, R., Kuriyan, J. & Marletta, M. A. Structural insights into the role of iron–histidine bond cleavage in nitric oxide-induced activation of H-NOX gas sensor proteins. Proc. Natl Acad. Sci. USA 111, E4156–E4164 (2014).
Plate, L. & Marletta, M. A. Nitric oxide-sensing H-NOX proteins govern bacterial communal behavior. Trends Biochem. Sci. 38, 566–575 (2013).
Zahn, J. A., Duncan, C. & DiSpirito, A. A. Oxidation of hydroxylamine by cytochrome P-460 of the obligate methylotroph Methylococcus capsulatus Bath. J. Bacteriol. 176, 5879–5887 (1994).
Luesken, F. A. et al. Simultaneous nitrite-dependent anaerobic methane and ammonium oxidation processes. Appl. Environ. Microbiol. 77, 6802–6807 (2011).
Beaumont, H. J. E., Lens, S. I., Westerhoff, H. V. & van Spanning, R. J. M. Novel nirK cluster genes in Nitrosomonas europaea are required for NirK-dependent tolerance to nitrite. J. Bacteriol. 187, 6849–6851 (2005).
Beaumont, H. J. E., van Schooten, B., Lens, S. I., Westerhoff, H. V. & van Spanning, R. J. M. Nitrosomonas europaea expresses a nitric oxide reductase during nitrification. J. Bacteriol. 186, 4417–4421 (2004).
Wrage, N., Velthof, G. L., van Beusichem, M. L. & Oenema, O. Role of nitrifier denitrification in the production of nitrous oxide. Soil Biol. Biochem. 33, 1723–1732 (2001).
Meincke, M., Bock, E., Kastrau, D. & Kroneck, P. M. H. Nitrite oxidoreductase from Nitrobacter hamburgensis: redox centers and their catalytic role. Arch. Microbiol. 158, 127–131 (1992).
Daims, H., Lücker, S. & Wagner, M. A new perspective on microbes formerly known as nitrite-oxidizing bacteria. Trends Microbiol. 24, 699–712 (2016).
Lehnert, N., Berto, T. C., Galinato, M. G. I. & Goodrich, L. E. in The Handbook of Porphyrin Science (eds Kadish, K. M., Smith, K. M. & Guilard, R.) 1–247 (World Scientific, New Jersey, 2011).
Ravishankara, A. R., Daniel, J. S. & Portmann, R. W. Nitrous oxide (N2O): the dominant ozone-depleting substance emitted in the 21st century. Science 326, 123–125 (2009).
Torres, M. J. et al. Nitrous oxide metabolism in nitrate-reducing bacteria: physiology and regulatory mechanisms. Advances in Microbial Physiology (ed. Poole, R. K.) 38, 353–432 (Elsevier, 2016).
Speelman, A. L. & Lehnert, N. Heme versus non-heme iron-nitroxyl {FeN(H)O}8 complexes: electronic structure and biologically relevant reactivity. Acc. Chem. Res. 47, 1106–1116 (2014).
Laughlin, R. J. & Stevens, R. J. Evidence for fungal dominance of denitrification and codenitrification in a grassland soil. Soil Sci. Soc. Am. J. 66, 1540–1548 (2002).
Crenshaw, C. L., Lauber, C., Sinsabaugh, R. L. & Stavely, L. K. Fungal control of nitrous oxide production in semiarid grassland. Biogeochem. 87, 17–27 (2008).
Long, A., Heitman, J., Tobias, C., Philips, R. & Song, B. Co-occurring anammox, denitrification, and codenitrification in agricultural soils. Appl. Environ. Microbiol. 79, 168–176 (2013).
Wankel, S. D. et al. Evidence for fungal and chemodenitrification based N2O flux from nitrogen impacted coastal sediments. Nat. Commun. 8, 15595 (2017).
Lewicka-Szczebak, D., Augustin, J., Giesemann, A. & Well, R. Quantifying N2O reduction to N2 based on N2O isotopocules — validation with independent methods (helium incubation and 15N gas flux method). Biogeoscience 14, 711–732 (2017).
Rohe, L., Well, R. & Lewicka-Szczebak, D. Use of oxygen isotopes to differentiate between nitrous oxide produced by fungi or bacteria during denitrification. Rapid Commun. Mass Spectrom. 31, 1297–1312 (2017).
Caranto, J. D., Weitz, A., Hendrich, M. P. & Kurtz Jr., D. M. The nitric oxide reductase mechanism of a flavo–diiron protein: identification of active-site intermediates and products. J. Am. Chem. Soc. 136, 7981–7992 (2014).
Zheng, S. et al. The functional model complex [Fe2(BPMP)(OPr)(NO)2](BPh4)2 provides insight into the mechanism of flavodiiron NO reductases. J. Am. Chem. Soc. 135, 4902–4905 (2013).
Maeda, K. et al. N2O production, a widespread trait in fungi. Sci. Rep. 5, 9697 (2015).
Shoun, H. & Tanimoto, T. Denitrification by the fungus Fusarium oxysporum and involvement of cytochrome P-450 in the respiratory nitrite reduction. J. Biol. Chem. 266, 11078–11082 (1991).
Shoun, H., Sudo, Y., Seto, Y. & Beppu, T. Purification and properties of a cyotchrome P-450 of a fungus. Fusarium oxysporum. J. Biochem. 94, 1219–1229 (1983).
Nittler, M. P., Hocking-Murray, D., Foo, C. K. & Sil, A. Identification of Histoplasma capsulatum transcripts induced in response to reactive nitrogen species. Mol. Biol. Cell 16, 4792–4813 (2005).
Yang, H., Gandhi, H., Ostrom, N. E. & Hegg, E. L. Isotopic fractionation by a fungal P450 nitric oxide reductase during the production of N2O. Environ. Sci. Technol. 48, 10707–10715 (2014).
Usuda, K., Toritsuka, N., Matsuo, Y., Kim, D. H. & Shoun, H. Denitrification by the fungus Cylindrocarpon tonkinense: anaerobic cell growth and two isozyme forms of cytochrome P-450nor. Appl. Environ. Microbiol. 61, 883–889 (1995).
Tsuruta, S. et al. Denitrification by yeasts and occurrence of cytochrome P450nor in Trichosporon cutaneum. FEMS Microbiol. Lett. 168, 105–110 (1998).
Shoun, H., Kim, D.-H., Uchiyama, H. & Sugiyama, J. Denitrification by fungi. FEMS Microbiol. Lett. 94, 277–281 (1992).
McQuarters, A. B., Wolf, M. W., Hunt, A. P. & Lehnert, N. 1958–2014: after 56 years of research, cytochrome P450 reactivity finally explained. Angew. Chem. Int. Ed. 53, 4750–4752 (2014).
McQuarters, A. B., Wirgau, N. E. & Lehnert, N. Model complexes of key intermediates in fungal cytochrome P450 nitric oxide reductase (P450nor). Curr. Opin. Chem. Biol. 19, 82–89 (2014).
Shoun, H., Fushinobu, S., Jiang, L., Kim, S.-W. & Wakagi, T. Fungal denitrification and nitric oxide reductase cytochrome P450nor. Phil. Trans. R. Soc. B 367, 1186–1194 (2012).
Shiro, Y. et al. Spectroscopic and kinetic studies on reaction of cytochrome P450nor with nitric oxide. J. Biol. Chem. 270, 1617–1623 (1995).
Shimizu, H. et al. Proton delivery in NO reduction by fungal nitric-oxide reductase. Cryogenic crystallography, spectroscopy, and kinetics of ferric-NO complexes of wild-type and mutant enzymes. J. Biol. Chem. 275, 4816–4826 (2000).
Obayashi, E. et al. Unique binding of nitric oxide to ferric nitric oxide reductase from Fusarium oxysporum elucidated with infrared, resonance Raman, and X-ray absorption spectroscopies. J. Am. Chem. Soc. 119, 7807–7816 (1997).
Paulat, F. & Lehnert, N. Electronic structure of ferric heme nitrosyl complexes with thiolate coordination. Inorg. Chem. 46, 1547–1549 (2007).
Praneeth, V. K. K. et al. Electronic structure of six-coordinate iron(iii)–porphyrin NO adducts: the elusive iron(iii)–NO(radical) state and its influence on the properties of these complexes. J. Am. Chem. Soc. 130, 15288–15303 (2008).
Lehnert, N., Scheidt, W. R. & Wolf, M. W. Structure and bonding in heme–nitrosyl complexes and implications for biology. Struct. Bond. 154, 155–223 (2014).
Soldatova, A. V., Ibrahim, M., Olson, J. S., Czernuszewicz, R. S. & Spiro, T. G. New light on NO Bonding in Fe(iii) heme proteins from resonance Raman spectroscopy and DFT modeling. J. Am. Chem. Soc. 132, 4614–4625 (2010).
Enemark, J. H. & Feltham, R. D. Principles of structure, bonding, and reactivity for metal nitrosyl complexes. Coord. Chem. Rev. 13, 339–406 (1974).
Wang, Y. & Averill, B. A. Direct observation by FTIR spectroscopy of the ferrous heme−NO+ intermediate in reduction of nitrite by a dissimilatory heme cd 1 nitrite reductase. J. Am. Chem. Soc. 118, 3972–3973 (1996).
Oshima, R. et al. Structural evidence for direct hydride transfer from NADH to cytochrome P450nor. J. Mol. Biol. 342, 207–217 (2004).
Riplinger, C. & Neese, F. The reaction mechanism of cytochrome P450 NO reductase: a detailed quantum mechanics/molecular mechanics study. ChemPhysChem. 12, 3192–3203 (2011).
Krámos, B., Menyhárd, D. K. & Oláh, J. Direct hydride shift mechanism and stereoselectivity of P450nor confirmed by QM/MM calculations. J. Phys. Chem. B 116, 872–885 (2012).
Riplinger, C. et al. New insights into the nature of observable reaction intermediates in cytochrome P450 NO reductase by using a combination of spectroscopy and quantum mechanics/molecular mechanics calculations. Chem. Eur. J. 20, 1602–1614 (2014).
Obayashi, E., Takahashi, S. & Shiro, Y. Electronic structure of reaction intermediate of cytochrome P450nor in its nitric oxide reduction. J. Am. Chem. Soc. 120, 12964–12965 (1998).
Lehnert, N., Praneeth, V. K. K. & Paulat, F. Electronic structure of iron(ii)–porphyrin nitroxyl complexes: molecular mechanism of fungal nitric oxide reductase (P450nor). J. Comput. Chem. 27, 1338–1351 (2006).
Yosca, T. H. et al. Iron(iv) hydroxide pK a and the role of thiolate ligation in C–H bond activation by cytochrome P450. Science 342, 825–829 (2013).
Yosca, T. H. et al. Setting an upper limit on the myoglobin iron(iv) hydroxide pK a: insight into axial ligand tuning in heme protein catalysis. J. Am. Chem. Soc. 136, 9124–9131 (2014).
Lin, R. & Farmer, P. J. The HNO adduct of myoglobin: synthesis and characterization. J. Am. Chem. Soc. 122, 2393–2394 (2000).
Lin, Y.-W. Rational design of metalloenzymes: from single to multiple active sites. Coord. Chem. Rev. 336, 1–27 (2017).
Wasser, I. M., de Vries, S., Moënne-Loccoz, P., Schröder, I. & Karlin, K. D. Nitric oxide in biological denitrification: Fe/Cu metalloenzymes and metal complex NOx redox chemistry. Chem. Rev. 102, 1201–1234 (2002).
Zumft, W. G. Nitric oxide reductases of prokaryotes with emphasis on the respiratory, heme–copper oxidase type. J. Inorg. Biochem. 99, 194–215 (2005).
Saraste, M. & Castresana, J. Cytochrome oxidase evolved by tinkering with denitrification enzymes. FEBS Lett. 341, 1–4 (1994).
Shiro, Y. Structure and function of bacterial nitric oxide reductases. Biochim. Biophys. Acta, Bioenerg. 1817, 1907–1913 (2012).
Hino, T. et al. Structural basis of biological N2O generation by bacterial nitric oxide reductase. Science 330, 1666–1670 (2010).
Timóteo, C. G. et al. Low-spin heme b 3 in the catalytic center of nitric oxide reductase from Pseudomonas nautica. Biochemistry 50, 4251–4262 (2011).
Blomberg, M. R. How quantum chemistry can solve fundamental problems in bioenergetics. Int. J. Quantum Chem. 115, 1197–1201 (2015).
Al-Attar, S. & de Vries, S. An electrogenic nitric oxide reductase. FEBS Lett. 589, 2050–2057 (2015).
Gonska, N. et al. Characterization of the quinol-dependent nitric oxide reductase from the pathogen Neisseria meningitidis, an electrogenic enzyme. Sci. Rep. 8, 3637 (2018).
Richardson, D., Felgate, H., Watmough, N., Thomson, A. & Baggs, E. Mitigating release of the potent greenhouse gas N2O from the nitrogen cycle — could enzymic regulation hold the key? Trends Biotechnol. 27, 388–397 (2009).
Kakishima, K., Shiratsuchi, A., Taoka, A., Nakanishi, Y. & Fukumori, Y. Participation of nitric oxide reductase in survival of Pseudomonas aeruginosa in LPS-activated macrophages. Biochem. Biophys. Res. Commun. 355, 587–591 (2007).
Yeung, N. et al. Rational design of a structural and functional nitric oxide reductase. Nature 462, 1079–1082 (2009).
Butler, C. S., Seward, H. E., Greenwood, C. & Thomson, A. J. Fast cytochrome bo from Escherichia coli binds two molecules of nitric oxide at CuB. Biochemistry 36, 16259–16266 (1997).
Girsch, P. & de Vries, S. Purification and initial kinetic and spectroscopic characterization of NO reductase from Paracoccus denitrificans. Biochim. Biophys. Acta, Bioenergy 1318, 202–216 (1997).
Watmough, N. J. et al. The dinuclear center of cytochrome bo 3 from Escherichia coli. J. Bioenerg. Biomembr. 30, 55–62 (1998).
Hendriks, J. et al. The active site of the bacterial nitric oxide reductase is a dinuclear iron center. Biochemistry 37, 13102–13109 (1998).
Moënne-Loccoz, P. & de Vries, S. Structural characterization of the catalytic high-spin heme b of nitric oxide reductase: a resonance Raman study. J. Am. Chem. Soc. 120, 5147–5152 (1998).
Cheesman, M. R., Zumft, W. G. & Thomson, A. J. The MCD and EPR of the heme centers of nitric oxide reductase from Pseudomonas stutzeri: evidence that the enzyme is structurally related to the heme–copper oxidases. Biochemistry 37, 3994–4000 (1998).
Flock, U., Reimann, J. & Ädelroth, P. Proton transfer in bacterial nitric oxide reductase. Biochem. Soc. Trans. 34, 188–190 (2006).
Speelman, A. L. et al. Unusual synthetic pathway for an {Fe(NO)2}9 dinitrosyl iron complex (DNIC) and insight into DNIC electronic structure via nuclear resonance vibrational spectroscopy. Inorg. Chem. 55, 5485–5501 (2016).
Praneeth, V. K. K., Näther, C., Peters, G. & Lehnert, N. Spectroscopic properties and electronic structure of five- and six-coordinate iron(ii) porphyrin NO complexes: effect of the axial N-donor ligand. Inorg. Chem. 45, 2795–2811 (2006).
Berto, T. C. et al. Characterization of the bridged hyponitrite complex {[Fe(OEP)]2(μ-N2O2)}: reactivity of hyponitrite complexes and biological relevance. Inorg. Chem. 53, 6398–6414 (2014).
Blomberg, M. R. A. Can reduction of NO to N2O in cytochrome c dependent nitric Oxide reductase proceed through a trans-mechanism? Biochemistry 56, 120–131 (2017).
Matsumura, H., Hayashi, T., Chakraborty, S., Lu, Y. & Moënne-Loccoz, P. The production of nitrous oxide by the heme/nonheme diiron center of engineered myoglobins (FeBMbs) proceeds through a trans-iron-nitrosyl dimer. J. Am. Chem. Soc. 136, 2420–2431 (2014).
Matsumura, H., Chakraborty, S., Reed, J., Lu, Y. & Moënne-Loccoz, P. Effect of outer-sphere side chain substitutions on the fate of the trans iron–nitrosyl dimer in heme/nonheme engineered myoglobins (FeBMbs): insights into the mechanism of denitrifying NO reductases. Biochemistry 55, 2091–2099 (2016).
Bhagi-Damodaran, A., Petrik, I. & Lu, Y. Using biosynthetic models of heme-copper oxidase and nitric oxide reductase in myoglobin to elucidate structural features responsible for enzymatic activities. Isr. J. Chem. 56, 773–790 (2016).
Chakraborty, S. et al. Recent advances in biosynthetic modeling of nitric oxide reductases and insights gained from nuclear resonance vibrational and other spectroscopic studies. Inorg. Chem. 54, 9317–9329 (2015).
Collman, J. P. et al. A functional nitric oxide reductase model. Proc. Natl Acad. Sci. USA 105, 15660–15665 (2008).
Blomberg, M. R. A. & Siegbahn, P. E. M. Mechanism for N2O generation in bacterial nitric oxide reductase: a quantum chemical study. Biochemistry 51, 5173–5186 (2012).
Blomberg, L. M., Blomberg, M. R. A. & Siegbahn, P. E. M. Reduction of nitric oxide in bacterial nitric oxide reductase — a theoretical model study. Biochim. Biophys. Acta, Bioenerg. 1757, 240–252 (2006).
Ye, R. W., Averill, B. A. & Tiedje, J. M. Denitrification: production and consumption of nitric oxide. Appl. Environ. Microbiol. 60, 1053–1058 (1994).
Hayashi, T. et al. Spectroscopic characterization of mononitrosyl complexes in heme–nonheme diiron centers within the myoglobin scaffold (FeBMbs): relevance to denitrifying NO reductase. Biochemistry 50, 5939–5947 (2011).
Tolman, W. B. Binding and activation of N2O at transition-metal centers: recent mechanistic insights. Angew. Chem. Int. Ed. 49, 1018–1024 (2010).
Severin, K. Synthetic chemistry with nitrous oxide. Chem. Soc. Rev. 44, 6375–6386 (2015).
Parmon, V. N., Panov, G. I., Uriarte, A. & Noskov, A. S. Nitrous oxide in oxidation chemistry and catalysis: application and production. Catal. Today 100, 115–131 (2005).
Gorelsky, S. I., Ghosh, S. & Solomon, E. I. Mechanism of N2O reduction by the μ4-S tetranuclear CuZ cluster of nitrous oxide reductase. J. Am. Chem. Soc. 128, 278–290 (2006).
Zeng, R., Feller, M., Ben-David, Y. & Milstein, D. Hydrogenation and hydrosilylation of nitrous oxide homogeneously catalyzed by a metal complex. J. Am. Chem. Soc. 139, 5720–5723 (2017).
Leont’ev, A. V., Fomicheva, O. A., Proskurnina, V. M. V. & Zefirov, S. N. S. Modern chemistry of nitrous oxide. Russ. Chem. Rev. 70, 91–104 (2001).
Armor, J. N. & Taube, H. Formation and reactions of [(NH3)5RuN2O2+]. J. Am. Chem. Soc. 91, 6874–6876 (1969).
Diamantis, A. A. & Sparrow, G. J. Nitrous oxide complexes: the isolation of pentammine(dinitrogen oxide)ruthenium(ii) tetrafluoroborate. Chem. Commun. 819–820 (1970).
Pamplin, C. B., Ma, E. S. F., Safari, N., Rettig, S. J. & James, B. R. The nitrous oxide complex, RuCl2(η1-N2O)(P−N)(PPh3) (P−N = [o-(N,N-dimethylamino)phenyl]diphenylphosphine); low temperature conversion of N2O to N2 and O2. J. Am. Chem. Soc. 123, 8596–8597 (2001).
Paulat, F. et al. Spectroscopic properties and electronic structure of pentammineruthenium(ii) dinitrogen oxide and corresponding nitrosyl complexes: binding mode of N2O and reactivity. Inorg. Chem. 43, 6979–6994 (2004).
Piro, N. A., Lichterman, M. F., Harman, W. H. & Chang, C. J. A structurally characterized nitrous oxide complex of vanadium. J. Am. Chem. Soc. 133, 2108–2111 (2011).
Dell’Acqua, S., Pauleta, S. R., Moura, I. & Moura, J. J. G. The tetranuclear copper active site of nitrous oxide reductase: the CuZ center. J. Biol. Inorg. Chem. 16, 183–194 (2011).
Riester, J., Zumft, W. G. & Kroneck, P. M. H. Nitrous oxide reductase from Pseudomonas stutzeri. Eur. J. Biochem. 178, 751–762 (1989).
McEwan, A. G., Greenfield, A. J., Wetzstein, H. G., Jackson, J. B. & Ferguson, S. J. Nitrous oxide reduction by members of the family Rhodospirillaceae and the nitrous oxide reductase of Rhodopseudomonas capsulata. J. Bacteriol. 164, 823–830 (1985).
Hole, U. H. et al. Characterization of the membranous denitrification enzymes nitrite reductase (cytochrome cd 1) and copper-containing nitrous oxide reductase from Thiobacillus denitrificans. Arch. Microbiol. 165, 55–61 (1996).
Prudêncio, M. et al. Purification, characterization, and preliminary crystallographic study of copper-containing nitrous oxide reductase from Pseudomonas nautica 617. Biochemistry 39, 3899–3907 (2000).
Brown, K. et al. A novel type of catalytic copper cluster in nitrous oxide reductase. Nat. Struct. Biol. 7, 191–195 (2000).
Chen, P., Cabrito, I., Moura, J. J. G., Moura, I. & Solomon, E. I. Spectroscopic and electronic structure studies of the μ4-sulfide bridged tetranuclear CuZ cluster in N2O reductase: molecular insight into the catalytic mechanism. J. Am. Chem. Soc. 124, 10497–10507 (2002).
Johnston, E. M. et al. Determination of the active form of the tetranuclear copper sulfur cluster in nitrous oxide reductase. J. Am. Chem. Soc. 136, 614–617 (2014).
Pomowski, A., Zumft, W. G., Kroneck, P. M. H. & Einsle, O. N2O binding at a [4Cu:2S] copper–sulphur cluster in nitrous oxide reductase. Nature 477, 234–237 (2011).
Paraskevopoulos, K., Antonyuk, S. V., Sawers, R. G., Eady, R. R. & Hasnain, S. S. Insight into catalysis of nitrous oxide reductase from high-resolution structures of resting and inhibitor-bound enzyme from Achromobacter cycloclastes. J. Mol. Biol. 362, 55–65 (2006).
Bar-Nahum, I. et al. Reduction of nitrous oxide to dinitrogen by a mixed valent tricopper-disulfido cluster. J. Am. Chem. Soc. 131, 2812–2814 (2009).
Johnson, B. J., Antholine, W. E., Lindeman, S. V., Graham, M. J. & Mankad, N. P. A one-hole Cu4S cluster with N2O reductase activity: a structural and functional model for CuZ*. J. Am. Chem. Soc. 138, 13107–13110 (2016).
Esmieu, C. et al. N2O reduction at a dissymmetric {Cu2S}-containing mixed-valent center. Chem. Sci. 5, 4774–4784 (2014).
Acknowledgements
Work on P450nor models and related NO complexes by the Lehnert laboratory is supported by a grant from the National Science Foundation (CHE-1464696 to N.L.), which is gratefully acknowledged.
Author information
Authors and Affiliations
Contributions
All authors contributed equally to the preparation of this manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Lehnert, N., Dong, H.T., Harland, J.B. et al. Reversing nitrogen fixation. Nat Rev Chem 2, 278–289 (2018). https://doi.org/10.1038/s41570-018-0041-7
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41570-018-0041-7
This article is cited by
-
Elucidating electron transfer pathways in N2OR catalysis for mitigation of N2O emissions: a comprehensive review
Reviews in Environmental Science and Bio/Technology (2024)
-
Paenibacillus lentimorbus alleviates nutrient deficiency-induced stress in Zea mays by modulating root system architecture, auxin signaling, and metabolic pathways
Plant Cell Reports (2024)
-
Frankia-actinorhizal symbiosis: a non-chemical biological assemblage for enhanced plant growth, nodulation and reclamation of degraded soils
Symbiosis (2024)
-
Pt-modified Fe3O4 Supported on Ni Foam Nanocomposite for Electrocatalytic Nitrate Reduction to Ammonia
Electrocatalysis (2024)
-
Laser-induced nitrogen fixation
Nature Communications (2023)