Abstract
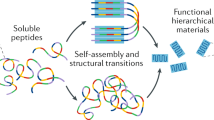
Nanostructures built from biomolecules such as proteins, DNA and RNA are attracting attention in many areas of biological and materials sciences. Such nanoscale engineering was pioneered with proteins, yet the use of DNA is rapidly gaining traction. What are the advantages of the different biopolymers and which is best suited for a given molecular structure, function or application? In this Review, we evaluate the different structural properties of proteins and nucleic acids, as well as possible designs and synthetic routes for functional nanostructures. By comparing protein engineering and DNA nanotechnology, we highlight molecular architectures that are relevant in biotechnology, biomedicine and synthetic biology research, and identify emerging areas for research such as hybrid materials composed of protein and DNA/RNA.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Jeschek, M. et al. Directed evolution of artificial metalloenzymes for in vivo metathesis. Nature 537, 661–665 (2016).
Renata, H., Wang, Z. J. & Arnold, F. H. Expanding the enzyme universe: accessing non-natural reactions by mechanism-guided directed evolution. Angew. Chem. Int. Ed. 54, 3351–3367 (2015).
Denard, C. A., Ren, H. & Zhao, H. Improving and repurposing biocatalysts via directed evolution. Curr. Opin. Chem. Biol. 25, 55–64 (2015).
Liszka, M. J., Clark, M. E., Schneider, E. & Clark, D. S. Nature versus nurture: developing enzymes that function under extreme conditions. Annu. Rev. Chem. Biomol. Eng. 3, 77–102 (2012).
Tiller, K. E. & Tessier, P. M. Advances in antibody design. Annu. Rev. Biomed. Eng. 17, 191–216 (2015).
Rosenbaum, L. Tragedy, perseverance, and chance — the story of CAR-T therapy. N. Engl. J. Med. 377, 1313–1315 (2017).
Rodriguez, E. A. et al. The growing and glowing toolbox of fluorescent and photoactive proteins. Trends Biochem. Sci. 42, 111–129 (2017).
Martell, J. D. et al. A split horseradish peroxidase for the detection of intercellular protein-protein interactions and sensitive visualization of synapses. Nat. Biotechnol. 34, 774–780 (2016).
Ellefson, J. W. et al. Synthetic evolutionary origin of a proofreading reverse transcriptase. Science 352, 1590–1593 (2016).
Seeman, N. C. Nanomaterials based on DNA. Annu. Rev. Biochem. 79, 65–87 (2010).
Rothemund, P. W. Folding DNA to create nanoscale shapes and patterns. Nature 440, 297–302 (2006).
Chen, Y. J., Groves, B., Muscat, R. A. & Seelig, G. DNA nanotechnology from the test tube to the cell. Nat. Nanotechnol. 10, 748–760 (2015).
Pinheiro, A. V., Han, D., Shih, W. M. & Yan, H. Challenges and opportunities for structural DNA nanotechnology. Nat. Nanotechnol. 6, 763–772 (2011).
Jones, M. R., Seeman, N. C. & Mirkin, C. A. Nanomaterials. Programmable materials and the nature of the DNA bond. Science 347, 1260901 (2015).
Seeman, N. C. & Sleiman, H. F. DNA nanotechnology. Nat. Rev. Mater. 3, 17068 (2017).
Torring, T., Voigt, N. V., Nangreave, J., Yan, H. & Gothelf, K. V. DNA origami: a quantum leap for self-assembly of complex structures. Chem. Soc. Rev. 40, 5636–5646 (2011).
Sacca, B. & Niemeyer, C. M. DNA origami: the art of folding DNA. Angew. Chem. Int. Ed. 51, 58–66 (2012).
Hong, F., Zhang, F., Liu, Y. & Yan, H. DNA origami: scaffolds for creating higher order structures. Chem. Rev. 117, 12584–12640 (2017).
Ellington, A. D. & Szostak, J. W. In vitro selection of RNA molecules that bind specific ligands. Nature 346, 818–822 (1990).
Tuerk, C. & Gold, L. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science 249, 505–510 (1990).
Zhou, J. & Rossi, J. Aptamers as targeted therapeutics: current potential and challenges. Nat. Rev. Drug. Discov. 16, 181–202 (2017).
Mor-Vaknin, N. et al. DEK-targeting DNA aptamers as therapeutics for inflammatory arthritis. Nat. Commun. 8, 14252 (2017).
Nimjee, S. M., White, R. R., Becker, R. C. & Sullenger, B. A. Aptamers as therapeutics. Annu. Rev. Pharmacol. Toxicol. 57, 61–79 (2017).
Gotrik, M. R., Feagin, T. A., Csordas, A. T., Nakamoto, M. A. & Soh, H. T. Advancements in aptamer discovery technologies. Acc. Chem. Res. 49, 1903–1910 (2016).
Attwood, T. K., Peffifer, S. R. & Thorne, D. Bioinformatics challenges at the interface of biology and computer science: Mind the gap. (John Wiley & Sons, 2016).
Dobson, C. M. Protein folding and misfolding. Nature 426, 884–890 (2003).
Kozlowski, L. P. Proteome-pI: proteome isoelectric point database. Nucleic Acids Res. 45, D1112–D1116 (2017).
Manning, G. S. The persistence length of DNA is reached from the persistence length of its null isomer through an internal electrostatic stretching force. Biophys. J. 91, 3607–3616 (2006).
Lane, M. D. & Seelig, B. Advances in the directed evolution of proteins. Curr. Opin. Chem. Biol. 22, 129–136 (2014).
Turner, N. J. Directed evolution drives the next generation of biocatalysts. Nat. Chem. Biol. 5, 567–573 (2009).
Packer, M. S. & Liu, D. R. Methods for the directed evolution of proteins. Nat. Rev. Genet. 16, 379–394 (2015).
Seelig, B. & Szostak, J. W. Selection and evolution of enzymes from a partially randomized non-catalytic scaffold. Nature 448, 828–831 (2007).
Wilson, D. S., Keefe, A. D. & Szostak, J. W. The use of mRNA display to select high-affinity protein-binding peptides. Proc. Natl Acad. Sci. USA 98, 3750–3755 (2001).
Boder, E. T. & Wittrup, K. D. Yeast surface display for screening combinatorial polypeptide libraries. Nat. Biotechnol. 15, 553–557 (1997).
Wang, X. X., Cho, Y. K. & Shusta, E. V. Mining a yeast library for brain endothelial cell-binding antibodies. Nat. Methods 4, 143–145 (2007).
Yi, L. et al. Engineering of TEV protease variants by yeast ER sequestration screening (YESS) of combinatorial libraries. Proc. Natl Acad. Sci. USA 110, 7229–7234 (2013).
Woolfson, D. N. et al. De novo protein design: how do we expand into the universe of possible protein structures? Curr. Opin. Struct. Biol. 33, 16–26 (2015).
Huang, P. S., Boyken, S. E. & Baker, D. The coming of age of de novo protein design. Nature 537, 320–327 (2016).
Damborsky, J. & Brezovsky, J. Computational tools for designing and engineering enzymes. Curr. Opin. Chem. Biol. 19, 8–16 (2014).
Hilvert, D. Design of protein catalysts. Annu. Rev. Biochem. 82, 447–470 (2013).
Khoury, G. A., Smadbeck, J., Kieslich, C. A. & Floudas, C. A. Protein folding and de novo protein design for biotechnological applications. Trends Biotechnol. 32, 99–109 (2014).
Rocklin, G. J. et al. Global analysis of protein folding using massively parallel design, synthesis, and testing. Science 357, 168–175 (2017).
Butterfield, G. L. et al. Evolution of a designed protein assembly encapsulating its own RNA genome. Nature 552, 415–420 (2017).
Chevalier, A. et al. Massively parallel de novo protein design for targeted therapeutics. Nature 550, 74–79 (2017).
Keefe, A. D., Pai, S. & Ellington, A. Aptamers as therapeutics. Nat. Rev. Drug. Discov. 9, 537–550 (2010).
Han, D. et al. Single-stranded DNA and RNA origami. Science 358, eaao2648 (2017).
Geary, C., Rothemund, P. W. & Andersen, E. S. RNA nanostructures. A single-stranded architecture for cotranscriptional folding of RNA nanostructures. Science 345, 799–804 (2014).
Wagenbauer, K. F., Sigl, C. & Dietz, H. Gigadalton-scale shape-programmable DNA assemblies. Nature 552, 78–83 (2017).
Tikhomirov, G., Petersen, P. & Qian, L. Fractal assembly of micrometre-scale DNA origami arrays with arbitrary patterns. Nature 552, 67–71 (2017).
Wickham, S. F. et al. A DNA-based molecular motor that can navigate a network of tracks. Nat. Nanotechnol. 7, 169–173 (2012).
Omabegho, T., Sha, R. & Seeman, N. C. A bipedal DNA Brownian motor with coordinated legs. Science 324, 67–71 (2009).
Derr, N. D. et al. Tug-of-war in motor protein ensembles revealed with a programmable DNA origami scaffold. Science 338, 662–665 (2012).
Wollman, A. J., Sanchez-Cano, C., Carstairs, H. M., Cross, R. A. & Turberfield, A. J. Transport and self-organization across different length scales powered by motor proteins and programmed by DNA. Nat. Nanotechnol. 9, 44–47 (2014).
Jung, C., Allen, P. B. & Ellington, A. D. A stochastic DNA walker that traverses a microparticle surface. Nat. Nanotechnol. 11, 157–163 (2016).
Li, J., Green, A. A., Yan, H. & Fan, C. Engineering nucleic acid structures for programmable molecular circuitry and intracellular biocomputation. Nat. Chem. 9, 1056–1067 (2017).
Amir, Y. et al. Universal computing by DNA origami robots in a living animal. Nat. Nanotechnol. 9, 353–357 (2014).
Burns, J. R., Seifert, A., Fertig, N. & Howorka, S. A biomimetic DNA-based channel for the ligand-controlled transport of charged molecular cargo across a biological membrane. Nat. Nanotechnol. 11, 152–156 (2016).
Langecker, M. et al. Synthetic lipid membrane channels formed by designed DNA nanostructures. Science 338, 932–936 (2012).
Bell, N. A. et al. DNA origami nanopores. Nano Lett. 12, 512–517 (2012).
Burns, J., Stulz, E. & Howorka, S. Self-assembled DNA nanopores that span lipid bilayers. Nano Lett. 13, 2351–2356 (2013).
Howorka, S. Building membrane nanopores. Nat. Nanotechnol. 12, 619–630 (2017).
Ayub, M. & Bayley, H. Engineered transmembrane pores. Curr. Opin. Chem. Biol. 34, 117–126 (2016).
Spiess, C. et al. Bispecific antibodies with natural architecture produced by co-culture of bacteria expressing two distinct half-antibodies. Nat. Biotechnol. 31, 753–758 (2013).
Brekke, O. H. & Sandlie, I. Therapeutic antibodies for human diseases at the dawn of the twenty-first century. Nat. Rev. Drug. Discov. 2, 52–62 (2003).
Kohler, G. & Milstein, C. Continuous cultures of fused cells secreting antibody of predefined specificity. Nature 256, 495–497 (1975).
Boulianne, G. L., Hozumi, N. & Shulman, M. J. Production of functional chimaeric mouse/human antibody. Nature 312, 643–646 (1984).
Better, M., Chang, C. P., Robinson, R. R. & Horwitz, A. H. Escherichia coli secretion of an active chimeric antibody fragment. Science 240, 1041–1043 (1988).
Jones, P. T., Dear, P. H., Foote, J., Neuberger, M. S. & Winter, G. Replacing the complementarity-determining regions in a human antibody with those from a mouse. Nature 321, 522–525 (1986).
de Wildt, R. M., Mundy, C. R., Gorick, B. D. & Tomlinson, I. M. Antibody arrays for high-throughput screening of antibody-antigen interactions. Nat. Biotechnol. 18, 989–994 (2000).
Kontermann, R. E. & Brinkmann, U. Bispecific antibodies. Drug Discov. Today 20, 838–847 (2015).
Bargou, R. et al. Tumor regression in cancer patients by very low doses of a T cell-engaging antibody. Science 321, 974–977 (2008).
Brinkmann, U. & Kontermann, R. E. The making of bispecific antibodies. MAbs 9, 182–212 (2017).
Wu, A. M. & Senter, P. D. Arming antibodies: prospects and challenges for immunoconjugates. Nat. Biotechnol. 23, 1137–1146 (2005).
Gross, G., Waks, T. & Eshhar, Z. Expression of immunoglobulin-T-cell receptor chimeric molecules as functional receptors with antibody-type specificity. Proc. Natl Acad. Sci. USA 86, 10024–10028 (1989).
Hamers-Casterman, C. et al. Naturally occurring antibodies devoid of light chains. Nature 363, 446–448 (1993).
Stanfield, R. L., Dooley, H., Flajnik, M. F. & Wilson, I. A. Crystal structure of a shark single-domain antibody V region in complex with lysozyme. Science 305, 1770–1773 (2004).
Muyldermans, S. Nanobodies: natural single-domain antibodies. Annu. Rev. Biochem. 82, 775–797 (2013).
Dolk, E. et al. Isolation of llama antibody fragments for prevention of dandruff by phage display in shampoo. Appl. Environ. Microbiol. 71, 442–450 (2005).
Hussack, G. et al. Neutralization of Clostridium difficile toxin A with single-domain antibodies targeting the cell receptor binding domain. J. Biol. Chem. 286, 8961–8976 (2011).
Rothbauer, U. et al. Targeting and tracing antigens in live cells with fluorescent nanobodies. Nat. Methods 3, 887–889 (2006).
Jost, C. & Pluckthun, A. Engineered proteins with desired specificity: DARPins, other alternative scaffolds and bispecific IgGs. Curr. Opin. Struct. Biol. 27, 102–112 (2014).
Binz, H. K. et al. High-affinity binders selected from designed ankyrin repeat protein libraries. Nat. Biotechnol. 22, 575–582 (2004).
Huber, T., Steiner, D., Rothlisberger, D. & Pluckthun, A. In vitro selection and characterization of DARPins and Fab fragments for the co-crystallization of membrane proteins: The Na+-citrate symporter CitS as an example. J. Struct. Biol. 159, 206–221 (2007).
Pluckthun, A. Designed ankyrin repeat proteins (DARPins): binding proteins for research, diagnostics, and therapy. Annu. Rev. Pharmacol. Toxicol. 55, 489–511 (2015).
Park, K. et al. Control of repeat-protein curvature by computational protein design. Nat. Struct. Mol. Biol. 22, 167–174 (2015).
Marold, J. D., Kavran, J. M., Bowman, G. D. & Barrick, D. A. Naturally occurring repeat protein with high internal sequence identity defines a new class of TPR-like proteins. Structure 23, 2055–2065 (2015).
Urvoas, A. et al. Design, production and molecular structure of a new family of artificial alpha-helicoidal repeat proteins (alphaRep) based on thermostable HEAT-like repeats. J. Mol. Biol. 404, 307–327 (2010).
Boersma, Y. L. & Pluckthun, A. DARPins and other repeat protein scaffolds: advances in engineering and applications. Curr. Opin. Biotechnol. 22, 849–857 (2011).
Parmeggiani, F. & Huang, P. S. Designing repeat proteins: a modular approach to protein design. Curr. Opin. Struct. Biol. 45, 116–123 (2017).
Driggers, E. M., Hale, S. P., Lee, J. & Terrett, N. K. The exploration of macrocycles for drug discovery — an underexploited structural class. Nat. Rev. Drug. Discov. 7, 608–624 (2008).
Hosseinzadeh, P. et al. Comprehensive computational design of ordered peptide macrocycles. Science 358, 1461–1466 (2017).
Ng, E. W. et al. Pegaptanib, a targeted anti-VEGF aptamer for ocular vascular disease. Nat. Rev. Drug. Discov. 5, 123–132 (2006).
Meng, H. M. et al. Aptamer-integrated DNA nanostructures for biosensing, bioimaging and cancer therapy. Chem. Soc. Rev. 45, 2583–2602 (2016).
Rinker, S., Ke, Y. G., Liu, Y., Chhabra, R. & Yan, H. Self-assembled DNA nanostructures for distance-dependent multivalent ligand-protein binding. Nat. Nanotechnol. 3, 418–422 (2008).
Fisher, M. A. & Tullman-Ercek, D. Change, exchange, and rearrange: protein engineering for the biotechnological production of fuels, pharmaceuticals, and other chemicals. Curr. Opin. Biotechnol. 24, 1010–1016 (2013).
Porter, J. L., Rusli, R. A. & Ollis, D. L. Directed evolution of enzymes for industrial biocatalysis. ChemBioChem 17, 197–203 (2016).
Bornscheuer, U. T. et al. Engineering the third wave of biocatalysis. Nature 485, 185–194 (2012).
Atsumi, S., Hanai, T. & Liao, J. C. Non-fermentative pathways for synthesis of branched-chain higher alcohols as biofuels. Nature 451, 86–89 (2008).
Steen, E. J. et al. Microbial production of fatty-acid-derived fuels and chemicals from plant biomass. Nature 463, 559–562 (2010).
Savile, C. K. et al. Biocatalytic asymmetric synthesis of chiral amines from ketones applied to sitagliptin manufacture. Science 329, 305–309 (2010).
O’Maille, P. E. et al. Quantitative exploration of the catalytic landscape separating divergent plant sesquiterpene synthases. Nat. Chem. Biol. 4, 617–623 (2008).
Lichman, B. R., Zhao, J., Hailes, H. C. & Ward, J. M. Enzyme catalysed Pictet-Spengler formation of chiral 1,1ʹ-disubstituted- and spiro-tetrahydroisoquinolines. Nat. Commun. 8, 14883 (2017).
Griengl, H., Schwab, H. & Fechter, M. The synthesis of chiral cyanohydrins by oxynitrilases. Trends Biotechnol. 18, 252–256 (2000).
DeSantis, G. et al. Creation of a productive, highly enantioselective nitrilase through gene site saturation mutagenesis (GSSM). J. Am. Chem. Soc. 125, 11476–11477 (2003).
Stemmer, W. P. Rapid evolution of a protein in vitro by DNA shuffling. Nature 370, 389–391 (1994).
Wells, J. A., Vasser, M. & Powers, D. B. Cassette mutagenesis: an efficient method for generation of multiple mutations at defined sites. Gene 34, 315–323 (1985).
Estell, D. A., Graycar, T. P. & Wells, J. A. Engineering an enzyme by site-directed mutagenesis to be resistant to chemical oxidation. J. Biol. Chem. 260, 6518–6521 (1985).
Prier, C. K., Zhang, R. K., Buller, A. R., Brinkmann-Chen, S. & Arnold, F. H. Enantioselective, intermolecular benzylic C–H amination catalysed by an engineered iron-haem enzyme. Nat. Chem. 9, 629–634 (2017).
Hammer, S. C. et al. Anti-Markovnikov alkene oxidation by metal-oxo-mediated enzyme catalysis. Science 358, 215–218 (2017).
Giger, L. et al. Evolution of a designed retro-aldolase leads to complete active site remodeling. Nat. Chem. Biol. 9, 494–498 (2013).
Chao, F. A. et al. Structure and dynamics of a primordial catalytic fold generated by in vitro evolution. Nat. Chem. Biol. 9, 81–83 (2013).
Dodani, S. C. et al. Discovery of a regioselectivity switch in nitrating P450s guided by molecular dynamics simulations and Markov models. Nat. Chem. 8, 419–425 (2016).
Chen, I., Dorr, B. M. & Liu, D. R. A general strategy for the evolution of bond-forming enzymes using yeast display. Proc. Natl Acad. Sci. USA 108, 11399–11404 (2011).
Preiswerk, N. et al. Impact of scaffold rigidity on the design and evolution of an artificial Diels–Alderase. Proc. Natl Acad. Sci. USA 111, 8013–8018 (2014).
Rothlisberger, D. et al. Kemp elimination catalysts by computational enzyme design. Nature 453, 190–195 (2008).
Kries, H., Blomberg, R. & Hilvert, D. De novo enzymes by computational design. Curr. Opin. Chem. Biol. 17, 221–228 (2013).
Lu, Y., Yeung, N., Sieracki, N. & Marshall, N. M. Design of functional metalloproteins. Nature 460, 855–862 (2009).
Keys, T. G. et al. Engineering the product profile of a polysialyltransferase. Nat. Chem. Biol. 10, 437–442 (2014).
Fox, R. J. et al. Improving catalytic function by ProSAR-driven enzyme evolution. Nat. Biotechnol. 25, 338–344 (2007).
Burton, A. J., Thomson, A. R., Dawson, W. M., Brady, R. L. & Woolfson, D. N. Installing hydrolytic activity into a completely de novo protein framework. Nat. Chem. 8, 837–844 (2016).
Blomberg, R. et al. Precision is essential for efficient catalysis in an evolved Kemp eliminase. Nature 503, 418–421 (2013).
Merski, M. & Shoichet, B. K. Engineering a model protein cavity to catalyze the Kemp elimination. Proc. Natl Acad. Sci. USA 109, 16179–16183 (2012).
Korendovych, I. V. et al. Design of a switchable eliminase. Proc. Natl Acad. Sci. USA 108, 6823–6827 (2011).
Bhan, N., Xu, P. & Koffas, M. A. Pathway and protein engineering approaches to produce novel and commodity small molecules. Curr. Opin. Biotechnol. 24, 1137–1143 (2013).
Kuchler, A., Yoshimoto, M., Luginbuhl, S., Mavelli, F. & Walde, P. Enzymatic reactions in confined environments. Nat. Nanotechnol. 11, 409–420 (2016).
Agapakis, C. M., Boyle, P. M. & Silver, P. A. Natural strategies for the spatial optimization of metabolism in synthetic biology. Nat. Chem. Biol. 8, 527–535 (2012).
Kell, D. B., Swainston, N., Pir, P. & Oliver, S. G. Membrane transporter engineering in industrial biotechnology and whole cell biocatalysis. Trends Biotechnol. 33, 237–246 (2015).
Cuenoud, B. & Szostak, J. W. A. DNA metalloenzyme with DNA ligase activity. Nature 375, 611–614 (1995).
Ponce-Salvatierra, A., Wawrzyniak-Turek, K., Steuerwald, U., Hobartner, C. & Pena, V. Crystal structure of a DNA catalyst. Nature 529, 231–234 (2016).
Horning, D. P. & Joyce, G. F. Amplification of RNA by an RNA polymerase ribozyme. Proc. Natl Acad. Sci. USA 113, 9786–9791 (2016).
Samanta, B. & Joyce, G. F. A reverse transcriptase ribozyme. eLife 6, e31153 (2017).
Cech, T. R. Structural biology. The ribosome is a ribozyme. Science 289, 878–879 (2000).
Ban, N., Nissen, P., Hansen, J., Moore, P. B. & Steitz, T. A. The complete atomic structure of the large ribosomal subunit at 2.4 A resolution. Science 289, 905–920 (2000).
Silverman, S. K. Catalytic DNA: scope, applications, and biochemistry of deoxyribozymes. Trends Biochem. Sci. 41, 595–609 (2016).
Pinheiro, V. B. et al. Synthetic genetic polymers capable of heredity and evolution. Science 336, 341–344 (2012).
Pinheiro, V. B. & Holliger, P. Towards XNA nanotechnology: new materials from synthetic genetic polymers. Trends Biotechnol. 32, 321–328 (2014).
Winnacker, M. & Kool, E. T. Artificial genetic sets composed of size-expanded base pairs. Angew. Chem. Int. Ed. 52, 12498–12508 (2013).
Rioz-Martinez, A. & Roelfes, G. DNA-based hybrid catalysis. Curr. Opin. Chem. Biol. 25, 80–87 (2015).
Boersma, A. J. et al. Catalytic enantioselective syn hydration of enones in water using a DNA-based catalyst. Nat. Chem. 2, 991–995 (2010).
Gartner, Z. J. et al. DNA-templated organic synthesis and selection of a library of macrocycles. Science 305, 1601–1605 (2004).
O’Reilly, R. K., Turberfield, A. J. & Wilks, T. R. The evolution of DNA-templated synthesis as a tool for materials discovery. Acc. Chem. Res. 50, 2496–2509 (2017).
Meng, W. et al. An autonomous molecular assembler for programmable chemical synthesis. Nat. Chem. 8, 542–548 (2016).
Alberts, B. et al. Molecular biology of the cell, Sixth edition. (Garland Publishing, 2014).
Sleytr, U. B., Schuster, B., Egelseer, E. M. & Pum, D. S-layers: principles and applications. FEMS Microbiol. Rev. 38, 823–864 (2014).
Fagan, R. P. & Fairweather, N. F. Biogenesis and functions of bacterial S-layers. Nat. Rev. Microbiol. 12, 211–222 (2014).
Missiakas, D. & Schneewind, O. Assembly and function of the Bacillus anthracis S-layer. Annu. Rev. Microbiol. 71, 79–98 (2017).
Baranova, E. et al. SbsB structure and lattice reconstruction unveil Ca2+ triggered S-layer assembly. Nature 487, 119–122 (2012).
van Raaij, M. J., Mitraki, A., Lavigne, G. & Cusack, S. A triple beta-spiral in the adenovirus fibre shaft reveals a new structural motif for a fibrous protein. Nature 401, 935–938 (1999).
Tanaka, S., Sawaya, M. R. & Yeates, T. O. Structure and mechanisms of a protein-based organelle in Escherichia coli. Science 327, 81–84 (2010).
Sutter, M., Greber, B., Aussignargues, C. & Kerfeld, C. A. Assembly principles and structure of a 6.5-MDa bacterial microcompartment shell. Science 356, 1293–1297 (2017).
Kerfeld, C. A., Heinhorst, S. & Cannon, G. C. Bacterial microcompartments. Annu. Rev. Microbiol. 64, 391–408 (2010).
Yeates, T. O., Crowley, C. S. & Tanaka, S. Bacterial microcompartment organelles: protein shell structure and evolution. Annu. Rev. Biophys. 39, 185–205 (2010).
McMahon, H. T. & Boucrot, E. Molecular mechanism and physiological functions of clathrin-mediated endocytosis. Nat. Rev. Mol. Cell Biol. 12, 517–533 (2011).
Peter, B. J. et al. BAR domains as sensors of membrane curvature: the amphiphysin BAR structure. Science 303, 495–499 (2004).
Johannes, L., Parton, R. G., Bassereau, P. & Mayor, S. Building endocytic pits without clathrin. Nat. Rev. Mol. Cell Biol. 16, 311–321 (2015).
Frost, A. et al. Structural basis of membrane invagination by F-BAR domains. Cell 132, 807–817 (2008).
Woolfson, D. N. & Mahmoud, Z. N. More than just bare scaffolds: towards multi-component and decorated fibrous biomaterials. Chem. Soc. Rev. 39, 3464–3479 (2010).
Ryadnov, M. G. & Woolfson, D. N. Engineering the morphology of a self-assembling protein fibre. Nat. Mater. 2, 329–332 (2003).
Burgess, N. C. et al. Modular design of self-assembling peptide-based nanotubes. J. Am. Chem. Soc. 137, 10554–10562 (2015).
Xu, C. et al. Rational design of helical nanotubes from self-assembly of coiled-coil lock washers. J. Am. Chem. Soc. 135, 15565–15578 (2013).
Magnotti, E. L. et al. Self-assembly of an alpha-helical peptide into a crystalline two-dimensional nanoporous framework. J. Am. Chem. Soc. 138, 16274–16282 (2016).
Brodin, J. D. et al. Metal-directed, chemically tunable assembly of one-, two- and three-dimensional crystalline protein arrays. Nat. Chem. 4, 375–382 (2012).
Suzuki, Y. et al. Self-assembly of coherently dynamic, auxetic, two-dimensional protein crystals. Nature 533, 369–373 (2016).
Gradisar, H. et al. Design of a single-chain polypeptide tetrahedron assembled from coiled-coil segments. Nat. Chem. Biol. 9, 362–366 (2013).
Padilla, J. E., Colovos, C. & Yeates, T. O. Nanohedra: using symmetry to design self assembling protein cages, layers, crystals, and filaments. Proc. Natl Acad. Sci. USA 98, 2217–2221 (2001).
Lai, Y. T. et al. Structure of a designed protein cage that self-assembles into a highly porous cube. Nat. Chem. 6, 1065–1071 (2014).
King, N. P. et al. Computational design of self-assembling protein nanomaterials with atomic level accuracy. Science 336, 1171–1174 (2012).
Mandell, D. J. & Kortemme, T. Computer-aided design of functional protein interactions. Nat. Chem. Biol. 5, 797–807 (2009).
Grueninger, D. et al. Designed protein–protein association. Science 319, 206–209 (2008).
Fletcher, J. M. et al. Self-assembling cages from coiled-coil peptide modules. Science 340, 595–599 (2013).
Kanekiyo, M. et al. Self-assembling influenza nanoparticle vaccines elicit broadly neutralizing H1N1 antibodies. Nature 499, 102–106 (2013).
Delpeyroux, F. et al. A poliovirus neutralization epitope expressed on hybrid hepatitis B surface antigen particles. Science 233, 472–475 (1986).
Schäffer, C. et al. Novel biocatalysts based on S-layer self-assembly of Geobacillus stearothermophilus NRS 2004/3a: a nanobiotechnological approach. Small 3, 1549–1559 (2007).
Ilk, N., Egelseer, E. M. & Sleytr, U. B. S-layer fusion proteins — construction principles and applications. Curr. Opin. Biotechnol. 22, 824–831 (2011).
Fan, C. et al. Short N-terminal sequences package proteins into bacterial microcompartments. Proc. Natl Acad. Sci. USA 107, 7509–7514 (2010).
Kotterman, M. A. & Schaffer, D. V. Engineering adeno-associated viruses for clinical gene therapy. Nat. Rev. Genet. 15, 445–451 (2014).
Shenton, W., Pum, D., Sleytr, U. B. & Mann, S. Synthesis of cadmium sulphide superlattices using self-assembled bacterial S-layers. Nature 389, 585–587 (1997).
Baneyx, F. & Matthaei, J. F. Self-assembled two-dimensional protein arrays in bionanotechnology: from S-layers to designed lattices. Curr. Opin. Biotechnol. 28, 39–45 (2014).
Howorka, S. Creating regular arrays of nanoparticles with self-assembling protein building blocks. J. Mater. Chem. 17, 2049–2053 (2007).
Nam, Y. S. et al. Biologically templated photocatalytic nanostructures for sustained light-driven water oxidation. Nat. Nanotechnol. 5, 340–344 (2010).
Lee, Y. J. et al. Fabricating genetically engineered high-power lithium-ion batteries using multiple virus genes. Science 324, 1051–1055 (2009).
Low, H. H. et al. Structure of a type IV secretion system. Nature 508, 550–553 (2014).
Winfree, E., Liu, F., Wenzler, L. A. & Seeman, N. C. Design and self-assembly of two-dimensional DNA crystals. Nature 394, 539–544 (1998).
Yan, H., Park, S. H., Finkelstein, G., Reif, J. H. & LaBean, T. H. DNA-templated self-assembly of protein arrays and highly conductive nanowires. Science 301, 1882–1884 (2003).
He, Y. et al. Hierarchical self-assembly of DNA into symmetric supramolecular polyhedra. Nature 452, 198–201 (2008).
Zheng, J. P. et al. From molecular to macroscopic via the rational design of a self-assembled 3D DNA crystal. Nature 461, 74–77 (2009).
Andersen, E. S. et al. Self-assembly of a nanoscale DNA box with a controllable lid. Nature 459, 73–76 (2009).
Douglas, S. M. et al. Self-assembly of DNA into nanoscale three-dimensional shapes. Nature 459, 414–418 (2009).
Castro, C. E. et al. A primer to scaffolded DNA origami. Nat. Methods 8, 221–229 (2011).
Douglas, S. M. et al. Rapid prototyping of 3D DNA-origami shapes with caDNAno. Nucleic Acids Res. 37, 5001–5006 (2009).
Benson, E. et al. DNA rendering of polyhedral meshes at the nanoscale. Nature 523, 441–444 (2015).
Veneziano, R. et al. Designer nanoscale DNA assemblies programmed from the top down. Science 352, 1534 (2016).
Dietz, H., Douglas, S. M. & Shih, W. M. Folding DNA into twisted and curved nanoscale shapes. Science 325, 725–730 (2009).
Han, D. et al. DNA origami with complex curvatures in three-dimensional space. Science 332, 342–346 (2011).
Kocabey, S. et al. Membrane-assisted growth of DNA origami nanostructure arrays. ACS Nano 9, 3530–3539 (2015).
Gerling, T., Wagenbauer, K. F., Neuner, A. M. & Dietz, H. Dynamic DNA devices and assemblies formed by shape-complementary, non-base pairing 3D components. Science 347, 1446–1452 (2015).
Woo, S. & Rothemund, P. W. Programmable molecular recognition based on the geometry of DNA nanostructures. Nat. Chem. 3, 620–627 (2011).
Goodman, R. P. et al. Rapid chiral assembly of rigid DNA building blocks for molecular nanofabrication. Science 310, 1661–1665 (2005).
Shih, W. M., Quispe, J. D. & Joyce, G. F. A. 1.7-kilobase single-stranded DNA that folds into a nanoscale octahedron. Nature 427, 618–621 (2004).
Zhang, F. et al. Complex wireframe DNA origami nanostructures with multi-arm junction vertices. Nat. Nanotechnol. 10, 779–784 (2015).
Wei, B., Dai, M. & Yin, P. Complex shapes self-assembled from single-stranded DNA tiles. Nature 485, 623–626 (2012).
Ke, Y., Ong, L. L., Shih, W. M. & Yin, P. Three-dimensional structures self-assembled from DNA bricks. Science 338, 1177–1183 (2012).
Zhang, Z. et al. Placing and shaping liposomes with reconfigurable DNA nanocages. Nat. Chem. 9, 653–659 (2017).
Howorka, S. Nanotechnology. Changing of the guard. Science 352, 890–891 (2016).
Franquelim, H. G., Khmelinskaia, A., Sobczak, J. P., Dietz, H. & Schwille, P. Membrane sculpting by curved DNA origami scaffolds. Nat. Commun. 9, 811 (2018).
Yoshina-Ishii, C. & Boxer, S. G. Arrays of mobile tethered vesicles on supported lipid bilayers. J. Am. Chem. Soc. 125, 3696–3697 (2003).
Baldelli Bombelli, F. et al. Closed nanoconstructs assembled by step-by-step ss-DNA coupling assisted by phospholipid membranes. Soft Matter 5, 1639–1645 (2009).
Johnson-Buck, A., Jiang, S., Yan, H. & Walter, N. G. DNA-cholesterol barges as programmable membrane-exploring agents. ACS Nano 8, 5641–5649 (2014).
Suzuki, Y., Endo, M. & Sugiyama, H. Lipid-bilayer-assisted two-dimensional self-assembly of DNA origami nanostructures. Nat. Commun. 6, 8052 (2015).
List, J., Weber, M. & Simmel, F. C. Hydrophobic actuation of a DNA origami bilayer structure. Angew. Chem. Int. Ed. 53, 4236–4239 (2014).
Czogalla, A. et al. Amphipathic DNA origami nanoparticles to scaffold and deform lipid membrane vesicles. Angew. Chem. Int. Ed. 54, 6501–6505 (2015).
Edwardson, T. G., Carneiro, K. M., McLaughlin, C. K., Serpell, C. J. & Sleiman, H. F. Site-specific positioning of dendritic alkyl chains on DNA cages enables their geometry-dependent self-assembly. Nat. Chem. 5, 868–875 (2013).
Yang, Y. et al. Self-assembly of size-controlled liposomes on DNA nanotemplates. Nat. Chem. 8, 476–483 (2016).
Xu, W. et al. A programmable DNA origami platform to organize SNAREs for membrane fusion. J. Am. Chem. Soc. 138, 4439–4447 (2016).
Perrault, S. D. & Shih, W. M. Virus-inspired membrane encapsulation of DNA nanostructures to achieve in vivo stability. ACS Nano 8, 5132–5140 (2014).
Howorka, S. DNA nanotechnology: bringing lipid bilayers into shape. Nat. Chem. 9, 611–613 (2017).
Jungmann, R. et al. Single-molecule kinetics and super-resolution microscopy by fluorescence imaging of transient binding on DNA origami. Nano Lett. 10, 4756–4761 (2010).
Iinuma, R. et al. Polyhedra self-assembled from DNA tripods and characterized with 3D DNA-PAINT. Science 344, 65–69 (2014).
Modi, S. et al. A DNA nanomachine that maps spatial and temporal pH changes inside living cells. Nat. Nanotechnol. 4, 325–330 (2009).
Modi, S., Nizak, C., Surana, S., Halder, S. & Krishnan, Y. Two DNA nanomachines map pH changes along intersecting endocytic pathways inside the same cell. Nat. Nanotechnol. 8, 459–467 (2013).
Knudsen, J. B. et al. Routing of individual polymers in designed patterns. Nat. Nanotechnol. 10, 892–898 (2015).
Tan, S. J., Campolongo, M. J., Luo, D. & Cheng, W. Building plasmonic nanostructures with DNA. Nat. Nanotechnol. 6, 268–276 (2011).
Ozbay, E. Plasmonics: merging photonics and electronics at nanoscale dimensions. Science 311, 189–193 (2006).
Klinkova, A., Choueiri, R. M. & Kumacheva, E. Self-assembled plasmonic nanostructures. Chem. Soc. Rev. 43, 3976–3991 (2014).
Mirkin, C. A., Letsinger, R. L., Mucic, R. C. & Storhoff, J. J. A DNA-based method for rationally assembling nanoparticles into macroscopic materials. Nature 382, 607–609 (1996).
Alivisatos, A. P. et al. Organization of ‘nanocrystal molecules’ using DNA. Nature 382, 609–611 (1996).
Zheng, J. W. et al. Two-dimensional nanoparticle arrays show the organizational power of robust DNA motifs. Nano Lett. 6, 1502–1504 (2006).
Aldaye, F. A., Palmer, A. L. & Sleiman, H. F. Assembling materials with DNA as the guide. Science 321, 1795–1799 (2008).
Sharma, J. et al. Control of self-assembly of DNA tubules through integration of gold nanoparticles. Science 323, 112–116 (2009).
Kuzyk, A. et al. DNA-based self-assembly of chiral plasmonic nanostructures with tailored optical response. Nature 483, 311–314 (2012).
Acuna, G. P. et al. Fluorescence enhancement at docking sites of DNA-directed self-assembled nanoantennas. Science 338, 506–510 (2012).
Sun, W. et al. Casting inorganic structures with DNA molds. Science 346, 1258361 (2014).
Trinh, T. et al. DNA-imprinted polymer nanoparticles with monodispersity and prescribed DNA-strand patterns. Nat. Chem. 10, 184–192 (2018).
Edwardson, T. G., Lau, K. L., Bousmail, D., Serpell, C. J. & Sleiman, H. F. Transfer of molecular recognition information from DNA nanostructures to gold nanoparticles. Nat. Chem. 8, 162–170 (2016).
Liu, W. et al. Diamond family of nanoparticle superlattices. Science 351, 582–586 (2016).
Kershner, R. J. et al. Placement and orientation of individual DNA shapes on lithographically patterned surfaces. Nat. Nanotechnol. 4, 557–561 (2009).
Hung, A. M. et al. Large-area spatially ordered arrays of gold nanoparticles directed by lithographically confined DNA origami. Nat. Nanotechnol. 5, 121–126 (2010).
Gopinath, A., Miyazono, E., Faraon, A. & Rothemund, P. W. Engineering and mapping nanocavity emission via precision placement of DNA origami. Nature 535, 401–405 (2016).
Lee, M. J. et al. Engineered synthetic scaffolds for organizing proteins within the bacterial cytoplasm. Nat. Chem. Biol. 14, 142–147 (2017).
Dueber, J. E. et al. Synthetic protein scaffolds provide modular control over metabolic flux. Nat. Biotechnol. 27, 753–759 (2009).
Flavell, R. R. & Muir, T. W. Expressed protein ligation (EPL) in the study of signal transduction, ion conduction, and chromatin biology. Acc. Chem. Res. 42, 107–116 (2009).
Dawson, P. E., Muir, T. W., Clark-Lewis, I. & Kent, S. B. Synthesis of proteins by native chemical ligation. Science 266, 776–779 (1994).
Chin, J. W. Expanding and reprogramming the genetic code. Nature 550, 53–60 (2017).
Spicer, C. D. & Davis, B. G. Selective chemical protein modification. Nat. Commun. 5, 4740 (2014).
Zolot, R. S., Basu, S. & Million, R. P. Antibody-drug conjugates. Nat. Rev. Drug. Discov. 12, 259–260 (2013).
Chudasama, V., Maruani, A. & Caddick, S. Recent advances in the construction of antibody-drug conjugates. Nat. Chem. 8, 114–119 (2016).
Maruani, A. et al. A plug-and-play approach to antibody-based therapeutics via a chemoselective dual click strategy. Nat. Commun. 6, 6645 (2015).
Chen, T., Hongdilokkul, N., Liu, Z., Thirunavukarasu, D. & Romesberg, F. E. The expanding world of DNA and RNA. Curr. Opin. Chem. Biol. 34, 80–87 (2016).
Praetorius, F. et al. Biotechnological mass production of DNA origami. Nature 552, 84–87 (2017).
Agarwal, N. P., Matthies, M., Joffroy, B. & Schmidt, T. L. Structural transformation of wireframe DNA origami DNA polymerase assisted gap-filling. ACS Nano 12, 2546–2553 (2018).
Surana, S., Shenoy, A. R. & Krishnan, Y. Designing DNA nanodevices for compatibility with the immune system of higher organisms. Nat. Nanotechnol. 10, 741–747 (2015).
Lee, D. S., Qian, H., Tay, C. Y. & Leong, D. T. Cellular processing and destinies of artificial DNA nanostructures. Chem. Soc. Rev. 45, 4199–4225 (2016).
Bell, N. A. & Keyser, U. F. Digitally encoded DNA nanostructures for multiplexed, single-molecule protein sensing with nanopores. Nat. Nanotechnol. 11, 645–651 (2016).
Dunn, K. E. et al. Guiding the folding pathway of DNA origami. Nature 525, 82–86 (2015).
Sacca, B. et al. Orthogonal protein decoration of DNA origami. Angew. Chem. Int. Ed. 49, 9378–9383 (2010).
Wilner, O. I. et al. Enzyme cascades activated on topologically programmed DNA scaffolds. Nat. Nanotechnol. 4, 249–254 (2009).
Linko, V., Eerikainen, M. & Kostiainen, M. A. A modular DNA origami-based enzyme cascade nanoreactor. Chem. Commun. 51, 5351–5354 (2015).
Zhao, Z. et al. Nanocaged enzymes with enhanced catalytic activity and increased stability against protease digestion. Nat. Commun. 7, 10619 (2016).
Fu, J. et al. Multi-enzyme complexes on DNA scaffolds capable of substrate channelling with an artificial swinging arm. Nat. Nanotechnol. 9, 531–536 (2014).
Grossi, G., Dalgaard Ebbesen Jepsen, M., Kjems, J. & Andersen, E. S. Control of enzyme reactions by a reconfigurable DNA nanovault. Nat. Commun. 8, 992 (2017).
Delebecque, C. J., Lindner, A. B., Silver, P. A. & Aldaye, F. A. Organization of intracellular reactions with rationally designed RNA assemblies. Science 333, 470–474 (2011).
Angelin, A. et al. Multiscale origami structures as interface for cells. Angew. Chem. Int. Ed. 54, 15813–15817 (2015).
Praetorius, F. & Dietz, H. Self-assembly of genetically encoded DNA-protein hybrid nanoscale shapes. Science 355, eaam5488 (2017).
Schiffels, D., Szalai, V. A. & Liddle, J. A. Molecular precision at micrometer length scales: hierarchical assembly of DNA–protein nanostructures. ACS Nano 11, 6623–6629 (2017).
Henning-Knechtel, A., Knechtel, J. & Magzoub, M. DNA-assisted oligomerization of pore-forming toxin monomers into precisely-controlled protein channels. Nucleic Acids Res. 45, 12057–12068 (2017).
Spruijt, E., Tusk, S. E. & Bayley, H. DNA scaffolds support stable and uniform peptide nanopores. Nat. Nanotechnol. https://doi.org/10.1038/s41565-018-0139-6 (2018).
Zhao, Y. H., Abraham, M. H. & Zissimos, A. M. Fast calculation of van der Waals volume as a sum of atomic and bond contributions and its application to drug compounds. J. Org. Chem. 68, 7368–7373 (2003).
Choi, J. & Majima, T. Conformational changes of non-B DNA. Chem. Soc. Rev. 40, 5893–5909 (2011).
Hansel-Hertsch, R. et al. G-Quadruplex structures mark human regulatory chromatin. Nat. Genet. 48, 1267–1272 (2016).
Colin, P. Y., Zinchenko, A. & Hollfelder, F. Enzyme engineering in biomimetic compartments. Curr. Opin. Struct. Biol. 33, 42–51 (2015).
Fischlechner, M. et al. Evolution of enzyme catalysts caged in biomimetic gel-shell beads. Nat. Chem. 6, 791–796 (2014).
He, M. & Taussig, M. J. Eukaryotic ribosome display with in situ DNA recovery. Nat. Methods 4, 281–288 (2007).
Carlson, J. C., Badran, A. H., Guggiana-Nilo, D. A. & Liu, D. R. Negative selection and stringency modulation in phage-assisted continuous evolution. Nat. Chem. Biol. 10, 216–222 (2014).
Tizei, P. A., Csibra, E., Torres, L. & Pinheiro, V. B. Selection platforms for directed evolution in synthetic biology. Biochem. Soc. Trans 44, 1165–1175 (2016).
Currin, A., Swainston, N., Day, P. J. & Kell, D. B. Synthetic biology for the directed evolution of protein biocatalysts: navigating sequence space intelligently. Chem. Soc. Rev. 44, 1172–1239 (2015).
Davids, T., Schmidt, M., Bottcher, D. & Bornscheuer, U. T. Strategies for the discovery and engineering of enzymes for biocatalysis. Curr. Opin. Chem. Biol. 17, 215–220 (2013).
Frushicheva, M. P. et al. Computer aided enzyme design and catalytic concepts. Curr. Opin. Chem. Biol. 21, 56–62 (2014).
Durao, P. et al. Opposing effects of folding and assembly chaperones on evolvability of Rubisco. Nat. Chem. Biol. 11, 148–155 (2015).
Balchin, D., Hayer-Hartl, M. & Hartl, F. U. In vivo aspects of protein folding and quality control. Science 353, aac4354 (2016).
Farías-Rico, J. A., Schmidt, S. & Höcker, B. Evolutionary relationship of two ancient protein superfolds. Nat. Chem. Biol. 10, 710–715 (2014).
Höcker, B. Design of proteins from smaller fragments-learning from evolution. Curr. Opin. Struct. Biol. 27, 56–62 (2014).
Ramakers, B. E., van Hest, J. C. & Lowik, D. W. Molecular tools for the construction of peptide-based materials. Chem. Soc. Rev. 43, 2743–2756 (2014).
Matteucci, M. D. & Caruthers, M. H. Synthesis of deoxyoligonucleotides on a polymer support. J. Am. Chem. Soc. 103, 3185–3191 (1981).
Kukwikila, M., Gale, N., El-Sagheer, A. H., Brown, T. & Tavassoli, A. Assembly of a biocompatible triazole-linked gene by one-pot click-DNA ligation. Nat. Chem. 9, 1089–1098 (2017).
Gibson, D. G. et al. Complete chemical synthesis, assembly, and cloning of a Mycoplasma genitalium genome. Science 319, 1215–1220 (2008).
Gibson, D. G. et al. Creation of a bacterial cell controlled by a chemically synthesized genome. Science 329, 52–56 (2010).
Shivalingam, A. & Brown, T. Synthesis of chemically modified DNA. Biochem. Soc. Trans 44, 709–715 (2016).
Chen, Z., Lichtor, P. A., Berliner, A. P., Chen, J. C. & Liu, D. R. Evolution of sequence-defined highly functionalized nucleic acid polymers. Nat. Chem. 10, 420–427 (2018).
Harris, L. J., Larson, S. B., Hasel, K. W. & McPherson, A. Refined structure of an intact IgG2a monoclonal antibody. Biochemistry 36, 1581–1597 (1997).
Garces, F. et al. Structural evolution of glycan recognition by a family of potent HIV antibodies. Cell 159, 69–79 (2014).
Guillard, S. et al. Structural and functional characterization of a DARPin which inhibits Ras nucleotide exchange. Nat. Commun. 8, 16111 (2017).
Kato, K. et al. Structural basis for specific inhibition of Autotaxin by a DNA aptamer. Nat. Struct. Mol. Biol. 23, 395–401 (2016).
Jiang, L. et al. De novo computational design of retro-aldol enzymes. Science 319, 1387–1391 (2008).
Sleytr, U. B. et al. Nanobiotechnology with S-layer proteins as building blocks. Prog. Mol. Biol. Transl Sci. 103, 277–352 (2011).
Marvin, D. A., Hale, R. D., Nave, C. & Helmer-Citterich, M. Molecular models and structural comparisons of native and mutant class I filamentous bacteriophages Ff (fd, f1, M13), If1 and IKe. J. Mol. Biol. 235, 260–286 (1994).
Schluenzen, F. et al. Structure of functionally activated small ribosomal subunit at 3.3 angstroms resolution. Cell 102, 615–623 (2000).
Acknowledgements
Supported the EPSRC (EP/N009282/1), the BBSRC (BB/M025373/1, BB/N017331/1), the Leverhulme Trust (RPG-2017-015) and Oxford Nanopore Technologies. The authors thank Keith Fox, Birte Höcker, and Derek Woolfson for critically reading the manuscript and suggesting improvements, and Katya Ahmad for calculating the van der Waals volumes of amino acids and nucleotides, and suggesting improvements for Box 1.
Author information
Authors and Affiliations
Contributions
G.C.P. and S.H. researched literature and wrote the article. J.R.B. contributed to the discussion of the content and editing of the manuscript, and generated the illustrations.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Pugh, G.C., Burns, J.R. & Howorka, S. Comparing proteins and nucleic acids for next-generation biomolecular engineering. Nat Rev Chem 2, 113–130 (2018). https://doi.org/10.1038/s41570-018-0015-9
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41570-018-0015-9
This article is cited by
-
Creating complex protocells and prototissues using simple DNA building blocks
Nature Communications (2023)
-
Highly shape- and size-tunable membrane nanopores made with DNA
Nature Nanotechnology (2022)
-
Design, assembly, and characterization of membrane-spanning DNA nanopores
Nature Protocols (2021)
-
A programmable polymer library that enables the construction of stimuli-responsive nanocarriers containing logic gates
Nature Chemistry (2020)
-
Synthetic protein-conductive membrane nanopores built with DNA
Nature Communications (2019)