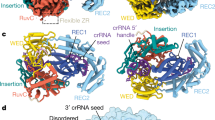
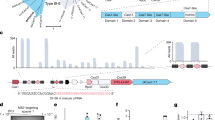
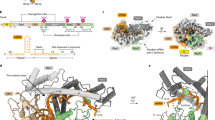
Abstract
RNA-guided binding and cleavage of nucleic acids by CRISPR–Cas systems is a defining feature of bacterial and archaeal adaptive immunity against viruses and plasmids. As a result of their programmable ability to cut specific DNA and RNA sequences, Cas9 and related single-subunit effector proteins from CRISPR–Cas systems have been widely adopted for research and therapeutic genome engineering applications. In this Review, we discuss the chemistry of macromolecules involved in the multistep interference pathway used by CRISPR–Cas systems that mediate accurate nucleic acid target recognition and cutting. Although this Review mainly focuses on DNA interference by Cas9, we briefly explore nucleic acid targeting by the single-effector proteins Cas12 and Cas13 to emphasize the conserved themes of precision DNA and RNA cleavage within class 2 CRISPR–Cas systems. We further highlight the unique mechanisms of surveillance complex formation, substrate recognition and target cleavage in molecular detail across diverse single-subunit CRISPR–Cas interference proteins.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Sorek, R., Kunin, V. & Hugenholtz, P. CRISPR — a widespread system that provides acquired resistance against phages in bacteria and archaea. Nat. Rev. Microbiol. 6, 181–186 (2008).
van der Oost, J., Westra, E. R., Jackson, R. N. & Wiedenheft, B. Unravelling the structural and mechanistic basis of CRISPR-Cas systems. Nat. Rev. Microbiol. 12, 479–492 (2014).
Makarova, K. S. et al. An updated evolutionary classification of CRISPR−Cas systems. Nat. Rev. Microbiol. 13, 722–736 (2015).
Mojica, F. J., Diez-Villasenor, C., Garcia-Martinez, J. & Soria, E. Intervening sequences of regularly spaced prokaryotic repeats derive from foreign genetic elements. J. Mol. Evol. 60, 174–182 (2005). This study uncovers the origin of spacers on the basis of homology to foreign DNA sequences and proposes that CRISPR is involved in specific immunity against MGEs.
Bolotin, A., Quinquis, B., Sorokin, A. & Ehrlich, S. D. Clustered regularly interspaced short palindrome repeats (CRISPRs) have spacers of extrachromosomal origin. Microbiology 151, 2551–2561 (2005).
Pourcel, C., Salvignol, G. & Vergnaud, G. CRISPR elements in Yersinia pestis acquire new repeats by preferential uptake of bacteriophage DNA, and provide additional tools for evolutionary studies. Microbiology 151, 653–663 (2005).
Barrangou, R. et al. CRISPR provides acquired resistance against viruses in prokaryotes. Science 315, 1709–1712 (2007). This is the first study to experimentally show the role of CRISPR systems in bacterial adaptive immunity.
Arslan, Z., Hermanns, V., Wurm, R., Wagner, R. & Pul, U. Detection and characterization of spacer integration intermediates in type IE CRISPR−Cas system. Nucleic Acids Res. 42, 7884–7893 (2014).
Nunez, J. K., Lee, A. S., Engelman, A. & Doudna, J. A. Integrase-mediated spacer acquisition during CRISPR−Cas adaptive immunity. Nature 519, 193–198 (2015).
Jansen, R., Embden, J. D., Gaastra, W. & Schouls, L. M. Identification of genes that are associated with DNA repeats in prokaryotes. Mol. Microbiol. 43, 1565–1575 (2002).
Kunin, V., Sorek, R. & Hugenholtz, P. Evolutionary conservation of sequence and secondary structures in CRISPR repeats. Genome Biol. 8, R61 (2007).
Brouns, S. J. et al. Small CRISPR RNAs guide antiviral defense in prokaryotes. Science 321, 960–964 (2008). This study shows that the formation of mature crRNAs containing the spacer and repeat sequences is essential for mediating an antiviral response.
Deltcheva, E. et al. CRISPR RNA maturation by trans-encoded small RNA and host factor RNase III. Nature 471, 602–607 (2011). This study identifies the tracrRNA molecule and shows its role in directing crRNA maturation and antiviral immunity.
Carte, J., Wang, R., Li, H., Terns, R. M. & Terns, M. P. Cas6 is an endoribonuclease that generates guide RNAs for invader defense in prokaryotes. Genes Dev. 22, 3489–3496 (2008).
Gesner, E. M., Schellenberg, M. J., Garside, E. L., George, M. M. & Macmillan, A. M. Recognition and maturation of effector RNAs in a CRISPR interference pathway. Nat. Struct. Mol. Biol. 18, 688–692 (2011).
Hochstrasser, M. L. & Doudna, J. A. Cutting it close: CRISPR-associated endoribonuclease structure and function. Trends Biochem. Sci. 40, 58–66 (2015).
Garneau, J. E. et al. The CRISPRCas bacterial immune system cleaves bacteriophage and plasmid DNA. Nature 468, 67–71 (2010).
Gasiunas, G., Barrangou, R., Horvath, P. & Siksnys, V. Cas9−crRNA ribonucleoprotein complex mediates specific DNA cleavage for adaptive immunity in bacteria. Proc. Natl Acad. Sci. USA 109, E2579–E2586 (2012).
Wiedenheft, B., Sternberg, S. H. & Doudna, J. A. RNA-guided genetic silencing systems in bacteria and archaea. Nature 482, 331–338 (2012).
Marraffini, L. A. & Sontheimer, E. J. CRISPR interference: RNA-directed adaptive immunity in bacteria and archaea. Nat. Rev. Genet. 11, 181–190 (2010).
Jinek, M. et al. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 337, 816–821 (2012). This study characterizes the determinants of DNA cleavage for SpCas9 and establishes programmable targeting using the crRNA–tracrRNA molecule or a chimeric sgRNA.
Cong, L. et al. Multiplex genome engineering using CRISPR–Cas systems. Science 339, 819–823 (2013).
Jinek, M. et al. RNA-programmed genome editing in human cells. eLife 2, e00471 (2013).
Mali, P. et al. RNA-guided human genome engineering via Cas9. Science 339, 823–826 (2013).
Rath, D., Amlinger, L., Hoekzema, M., Devulapally, P. R. & Lundgren, M. Efficient programmable gene silencing by Cascade. Nucleic Acids Res. 43, 237–246 (2015).
O’Connell, M. R. et al. Programmable RNA recognition and cleavage by CRISPR–Cas9. Nature 516, 263–266 (2014).
Gilbert, L. A. et al. Genome-scale CRISPR-mediated control of gene repression and activation. Cell 159, 647–661 (2014).
Larson, M. H. et al. CRISPR interference (CRISPRi) for sequence-specific control of gene expression. Nat. Protoc. 8, 2180–2196 (2013).
Doudna, J. A. & Charpentier, E. Genome editing. The new frontier of genome engineering with CRISPR-Cas9. Science 346, 1258096 (2014).
Hsu, P. D., Lander, E. S. & Zhang, F. Development and applications of CRISPR-Cas9 for genome engineering. Cell 157, 1262–1278 (2014).
Mali, P., Esvelt, K. M. & Church, G. M. Cas9 as a versatile tool for engineering biology. Nat. Methods 10, 957–963 (2013).
Sander, J. D. & Joung, J. K. CRISPR−Cas systems for editing, regulating and targeting genomes. Nat. Biotechnol. 32, 347–355 (2014).
Fellmann, C., Gowen, B. G., Lin, P. C., Doudna, J. A. & Corn, J. E. Cornerstones of CRISPR−Cas in drug discovery and therapy. Nat. Rev. Drug Discov. 16, 89–100 (2017).
Barrangou, R. & Horvath, P. A decade of discovery: CRISPR functions and applications. Nat. Microbiol. 2, 17092 (2017).
Burstein, D. et al. New CRISPRCas systems from uncultivated microbes. Nature 542, 237–241 (2017).
Shmakov, S. et al. Discovery and functional characterization of diverse class 2 CRISPR−Cas systems. Mol. Cell 60, 385–397 (2015). This study reports the discovery of three distinct class 2 CRISPR–Cas systems (C2c1, C2c2 and C2c3) and shows that these CRISPR loci contain functional interference proteins.
Shmakov, S. et al. Diversity and evolution of class 2 CRISPR−Cas systems. Nat. Rev. Microbiol. 15, 169–182 (2017). This review provides a comprehensive overview of all class 2 CRISPR–Cas systems discovered to date and discusses their evolutionary origins and relationships.
Stern, A. & Sorek, R. The phage-host arms race: shaping the evolution of microbes. Bioessays 33, 43–51 (2011).
Koonin, E. V. & Makarova, K. S. CRISPR−Cas: evolution of an RNA-based adaptive immunity system in prokaryotes. RNA Biol. 10, 679–686 (2013).
Zhang, J., Kasciukovic, T. & White, M. F. The CRISPR associated protein Cas4 is a 5′ to 3’ DNA exonuclease with an iron-sulfur cluster. PLoS ONE 7, e47232 (2012).
Nam, K. H., Kurinov, I. & Ke, A. Crystal structure of clustered regularly interspaced short palindromic repeats (CRISPR)-associated Csn2 protein revealed Ca2+-dependent double-stranded DNA binding activity. J. Biol. Chem. 286, 30759–30768 (2011).
Arslan, Z. et al. Double-strand DNA end-binding and sliding of the toroidal CRISPR-associated protein Csn2. Nucleic Acids Res. 41, 6347–6359 (2013).
Kazlauskiene, M., Kostiuk, G., Venclovas, C., Tamulaitis, G. & Siksnys, V. A cyclic oligonucleotide signaling pathway in type III CRISPR−Cas systems. Science 357, 605–609 (2017).
Niewoehner, O. et al. Type III CRISPR−Cas systems produce cyclic oligoadenylate second messengers. Nature 548, 543–548 (2017).
Mohanraju, P. et al. Diverse evolutionary roots and mechanistic variations of the CRISPR−Cas systems. Science 353, aad5147 (2016).
Koonin, E. V., Makarova, K. S. & Zhang, F. Diversity, classification and evolution of CRISPR−Cas systems. Curr. Opin. Microbiol. 37, 67–78 (2017).
Sinkunas, T. et al. Cas3 is a single-stranded DNA nuclease and ATP-dependent helicase in the CRISPR−Cas immune system. EMBO J. 30, 1335–1342 (2011).
Tamulaitis, G., Venclovas, C. & Siksnys, V. Type III CRISPR−Cas immunity: major differences brushed aside. Trends Microbiol. 25, 49–61 (2017).
Samai, P. et al. Co-transcriptional DNA and RNA cleavage during type III CRISPR−Cas immunity. Cell 161, 1164–1174 (2015).
Liu, T. Y., Iavarone, A. T. & Doudna, J. A. RNA and DNA targeting by a reconstituted thermus thermophilus type III-A CRISPR−Cas system. PLoS ONE 12, e0170552 (2017).
Elmore, J. R. et al. Bipartite recognition of target RNAs activates DNA cleavage by the Type IIIB CRISPR−Cas system. Genes Dev. 30, 447–459 (2016).
Estrella, M. A., Kuo, F. T. & Bailey, S. RNA-activated DNA cleavage by the Type IIIB CRISPR−Cas effector complex. Genes Dev. 30, 460–470 (2016).
Makarova, K. S., Wolf, Y. I. & Koonin, E. V. Comparative genomics of defense systems in archaea and bacteria. Nucleic Acids Res. 41, 4360–4377 (2013).
Makarova, K. S. et al. Evolution and classification of the CRISPR−Cas systems. Nat. Rev. Microbiol. 9, 467–477 (2011).
Chylinski, K., Makarova, K. S., Charpentier, E. & Koonin, E. V. Classification and evolution of type II CRISPRCas systems. Nucleic Acids Res. 42, 6091–6105 (2014).
Heler, R. et al. Cas9 specifies functional viral targets during CRISPR−Cas adaptation. Nature 519, 199–202 (2015).
Aravind, L., Makarova, K. S. & Koonin, E. V. Survey and summary: Holliday junction resolvases and related nucleases: identification of new families, phyletic distribution and evolutionary trajectories. Nucleic Acids Res. 28, 3417–3432 (2000).
Zetsche, B. et al. Cpf1 is a single RNA-guided endonuclease of a class 2 CRISPR−Cas system. Cell 163, 759–771 (2015).
Swarts, D. C., van der Oost, J. & Jinek, M. Structural basis for guide RNA processing and seed-dependent DNA targeting by CRISPR−Cas12a. Mol. Cell 66, 221–233.e4 (2017).
Fonfara, I., Richter, H., Bratovic, M., Le Rhun, A. & Charpentier, E. The CRISPR-associated DNA-cleaving enzyme Cpf1 also processes precursor CRISPR RNA. Nature 532, 517–521 (2016). This study shows the most streamlined CRISPR–Cas system by demonstrating that Cpf1 is independently capable of both crRNA processing and DNA target interference.
Anantharaman, V., Makarova, K. S., Burroughs, A. M., Koonin, E. V. & Aravind, L. Comprehensive analysis of the HEPN superfamily: identification of novel roles in intra-genomic conflicts, defense, pathogenesis and RNA processing. Biol. Direct 8, 15 (2013).
Smargon, A. A. et al. Cas13b is a type VIB CRISPR-associated RNA-guided RNase differentially regulated by accessory proteins Csx27 and Csx28. Mol. Cell 65, 618–630.e7 (2017).
Abudayyeh, O. O. et al. C2c2 is a single-component programmable RNA-guided RNA-targeting CRISPR effector. Science 353, aaf5573 (2016). This study validates a novel class of CRISPR interference proteins capable of programmable, RNA-guided RNA cleavage and uncovers its nonspecific RNase activity.
East-Seletsky, A. et al. Two distinct RNase activities of CRISPR−C2c2 enable guide-RNA processing and RNA detection. Nature 538, 270–273 (2016).
Liu, L. et al. Two distant catalytic sites are responsible for C2c2 RNase activities. Cell 168, 121–134.e12 (2017).
Liu, L. et al. The molecular architecture for RNA-guided RNA cleavage by Cas13a. Cell 170, 714–726.e10 (2017).
Karvelis, T. et al. crRNA and tracrRNA guide Cas9-mediated DNA interference in Streptococcus thermophilus. RNA Biol. 10, 841–851 (2013).
Chylinski, K., Le Rhun, A. & Charpentier, E. The tracrRNA and Cas9 families of type II CRISPR−Cas immunity systems. RNA Biol. 10, 726–737 (2013).
Yang, H., Gao, P., Rajashankar, K. R. & Patel, D. J. PAM-dependent target DNA recognition and cleavage by C2c1 CRISPR–Cas endonuclease. Cell 167, 1814–1828.e12 (2016).
Burstein, D. et al. New CRISPR–Cas systems from uncultivated microbes. Nature 542, 237–241 (2017).
Hsu, P. D. et al. DNA targeting specificity of RNA-guided Cas9 nucleases. Nat. Biotechnol. 31, 827–832 (2013).
Dang, Y. et al. Optimizing sgRNA structure to improve CRISPR–Cas9 knockout efficiency. Genome Biol. 16, 280 (2015).
Hendel, A. et al. Chemically modified guide RNAs enhance CRISPR-Cas genome editing in human primary cells. Nat. Biotechnol. 33, 985–989 (2015).
Fu, Y., Sander, J. D., Reyon, D., Cascio, V. M. & Joung, J. K. Improving CRISPR−Cas nuclease specificity using truncated guide RNAs. Nat. Biotechnol. 32, 279–284 (2014).
Nissim, L., Perli, S. D., Fridkin, A., Perez-Pinera, P. & Lu, T. K. Multiplexed and programmable regulation of gene networks with an integrated RNA and CRISPR−Cas toolkit in human cells. Mol. Cell 54, 698–710 (2014).
Doench, J. G. et al. Rational design of highly active sgRNAs for CRISPR–Cas9-mediated gene inactivation. Nat. Biotechnol. 32, 1262–1267 (2014).
Konermann, S. et al. Genome-scale transcriptional activation by an engineered CRISPR–Cas9 complex. Nature 517, 583–588 (2015).
Nowak, C. M., Lawson, S., Zerez, M. & Bleris, L. Guide RNA engineering for versatile Cas9 functionality. Nucleic Acids Res. 44, 9555–9564 (2016).
Chen, B. et al. Dynamic imaging of genomic loci in living human cells by an optimized CRISPR−Cas system. Cell 155, 1479–1491 (2013).
Thyme, S. B., Akhmetova, L., Montague, T. G., Valen, E. & Schier, A. F. Internal guide RNA interactions interfere with Cas9-mediated cleavage. Nat. Commun. 7, 11750 (2016).
Lim, Y. et al. Structural roles of guide RNAs in the nuclease activity of Cas9 endonuclease. Nat. Commun. 7, 13350 (2016).
Ma, E., Harrington, L. B., O’Connell, M. R., Zhou, K. & Doudna, J. A. Single-stranded DNA cleavage by divergent CRISPR–Cas9 enzymes. Mol. Cell 60, 398–407 (2015).
Jiang, F., Zhou, K., Ma, L., Gressel, S. & Doudna, J. A. Structural Biology. A Cas9-guide RNA complex preorganized for target DNA recognition. Science 348, 1477–1481 (2015).
Nishimasu, H. et al. Crystal structure of Cas9 in complex with guide RNA and target DNA. Cell 156, 935–949 (2014).
Anders, C., Niewoehner, O., Duerst, A. & Jinek, M. Structural basis of PAM-dependent target DNA recognition by the Cas9 endonuclease. Nature 513, 569–573 (2014).
Jiang, F. et al. Structures of a CRISPR–Cas9 R-loop complex primed for DNA cleavage. Science 351, 867–871 (2016).
Briner, A. E. et al. Guide RNA functional modules direct Cas9 activity and orthogonality. Mol. Cell 56, 333–339 (2014).
Wright, A. V. et al. Rational design of a split-Cas9 enzyme complex. Proc. Natl Acad. Sci. USA 112, 2984–2989 (2015).
Sternberg, S. H., LaFrance, B., Kaplan, M. & Doudna, J. A. Conformational control of DNA target cleavage by CRISPR–Cas9. Nature 527, 110–113 (2015). This study shows that the conformation of the HNH nuclease domain within SpCas9 regulates RuvC nuclease activity to ensure accurate and concerted cleavage of both DNA strands.
Nishimasu, H. et al. Crystal structure of Staphylococcus aureus Cas9. Cell 162, 1113–1126 (2015).
Hirano, H. et al. Structure and engineering of Francisella novicida Cas9. Cell 164, 950–961 (2016).
Esvelt, K. M. et al. Orthogonal Cas9 proteins for RNA-guided gene regulation and editing. Nat. Methods 10, 1116–1121 (2013).
Yamada, M. et al. Crystal structure of the minimal Cas9 from Campylobacter jejuni reveals the molecular diversity in the CRISPR-Cas9 systems. Mol. Cell 65, 1109–1121.e3 (2017).
Liu, L. et al. C2c1−sgRNA complex structure reveals RNA-guided DNA cleavage mechanism. Mol. Cell 65, 310–322 (2017).
Yang, H., Gao, P., Rajashankar, K. R. & Patel, D. J. PAM-dependent target DNA recognition and cleavage by C2c1 CRISPR−Cas endonuclease. Cell 167, 1814–1828.e12 (2016).
Dong, D. et al. The crystal structure of Cpf1 in complex with CRISPR RNA. Nature 532, 522–526 (2016).
Gao, P., Yang, H., Rajashankar, K. R., Huang, Z. & Patel, D. J. Type V CRISPR−Cas Cpf1 endonuclease employs a unique mechanism for crRNA-mediated target DNA recognition. Cell Res. 26, 901–913 (2016).
Yamano, T. et al. Crystal structure of Cpf1 in complex with guide RNA and target DNA. Cell 165, 949–962 (2016).
Stella, S., Alcon, P. & Montoya, G. Structure of the Cpf1 endonuclease R-loop complex after target DNA cleavage. Nature 546, 559–563 (2017).
Mojica, F. J., Diez-Villasenor, C., Garcia-Martinez, J. & Almendros, C. Short motif sequences determine the targets of the prokaryotic CRISPR defence system. Microbiology 155, 733–740 (2009).
Shah, S. A., Erdmann, S., Mojica, F. J. & Garrett, R. A. Protospacer recognition motifs: mixed identities and functional diversity. RNA Biol. 10, 891–899 (2013).
Deveau, H. et al. Phage response to CRISPR-encoded resistance in Streptococcus thermophilus. J. Bacteriol. 190, 1390–1400 (2008).
Sternberg, S. H., Redding, S., Jinek, M., Greene, E. C. & Doudna, J. A. DNA interrogation by the CRISPR RNA-guided endonuclease Cas9. Nature 507, 62–67 (2014).
Tsai, S. Q. et al. GUIDE-seq enables genome-wide profiling of off-target cleavage by CRISPR−Cas nucleases. Nat. Biotechnol. 33, 187–197 (2015).
Jiang, W., Bikard, D., Cox, D., Zhang, F. & Marraffini, L. A. RNA-guided editing of bacterial genomes using CRISPR−Cas systems. Nat. Biotechnol. 31, 233–239 (2013).
Westra, E. R. et al. CRISPR immunity relies on the consecutive binding and degradation of negatively supercoiled invader DNA by Cascade and Cas3. Mol. Cell 46, 595–605 (2012).
Knight, S. C. et al. Dynamics of CRISPR–Cas9 genome interrogation in living cells. Science 350, 823–826 (2015).
Singh, D., Sternberg, S. H., Fei, J., Doudna, J. A. & Ha, T. Real-time observation of DNA recognition and rejection by the RNA-guided endonuclease Cas9. Nat. Commun. 7, 12778 (2016).
Hirano, S., Nishimasu, H., Ishitani, R. & Nureki, O. Structural basis for the altered PAM specificities of engineered CRISPR–Cas9. Mol. Cell 61, 886–894 (2016).
Kleinstiver, B. P. et al. Engineered CRISPR–Cas9 nucleases with altered PAM specificities. Nature 523, 481–485 (2015).
Mekler, V., Minakhin, L. & Severinov, K. Mechanism of duplex DNA destabilization by RNA-guided Cas9 nuclease during target interrogation. Proc. Natl Acad. Sci. USA 114, 5443–5448 (2017).
Gao, L. et al. Engineered Cpf1 variants with altered PAM specificities. Nat. Biotechnol. 35, 789–792 (2017).
Nishimasu, H. et al. Structural basis for the altered PAM recognition by engineered CRISPR−Cpf1. Mol. Cell 67, 139–147.e2 (2017).
Yamano, T. et al. Structural basis for the canonical and non-canonical PAM recognition by CRISPR−Cpf1. Mol. Cell. 67, 633–645.e3 (2017).
Jore, M. M. et al. Structural basis for CRISPR RNA-guided DNA recognition by Cascade. Nat. Struct. Mol. Biol. 18, 529–536 (2011).
Szczelkun, M. D. et al. Direct observation of R-loop formation by single RNA-guided Cas9 and Cascade effector complexes. Proc. Natl Acad. Sci. USA 111, 9798–9803 (2014).
Rutkauskas, M. et al. Directional R-loop formation by the CRISPR−Cas surveillance complex cascade provides efficient off-target site rejection. Cell Rep. 10, 1534–1543 (2015).
Pattanayak, V. et al. High-throughput profiling of off-target DNA cleavage reveals RNA-programmed Cas9 nuclease specificity. Nat. Biotechnol. 31, 839–843 (2013).
Wu, X. et al. Genome-wide binding of the CRISPR endonuclease Cas9 in mammalian cells. Nat. Biotechnol. 32, 670–676 (2014).
Zheng, T. et al. Profiling single-guide RNA specificity reveals a mismatch sensitive core sequence. Sci. Rep. 7, 40638 (2017).
Coulombe, B. & Burton, Z. F. DNA bending and wrapping around RNA polymerase: a “revolutionary” model describing transcriptional mechanisms. Microbiol. Mol. Biol. Rev. 63, 457–478 (1999).
Westra, E. R. et al. Cascade-mediated binding and bending of negatively supercoiled DNA. RNA Biol. 9, 1134–1138 (2012).
Hochstrasser, M. L., Taylor, D. W., Kornfeld, J. E., Nogales, E. & Doudna, J. A. DNA targeting by a minimal CRISPR RNA-guided cascade. Mol. Cell 63, 840–851 (2016).
Xiao, Y. et al. Structure basis for directional R-loop formation and substrate handover mechanisms in type I CRISPR−Cas system. Cell 170, 48–60.e11 (2017).
Nelson, P. Transport of torsional stress in DNA. Proc. Natl Acad. Sci. USA 96, 14342–14347 (1999).
Vassylyev, D. G. et al. Crystal structure of a bacterial RNA polymerase holoenzyme at 2.6 Å resolution. Nature 417, 712–719 (2002).
Josephs, E. A. et al. Structure and specificity of the RNA-guided endonuclease Cas9 during DNA interrogation, target binding and cleavage. Nucleic Acids Res. 43, 8924–8941 (2015).
Farasat, I. & Salis, H. M. A biophysical model of CRISPR–Cas9 activity for rational design of genome editing and gene regulation. PLoS Comput. Biol. 12, e1004724 (2016).
Cencic, R. et al. Protospacer adjacent motif (PAM)-distal sequences engage CRISPR–Cas9 DNA target cleavage. PLoS ONE 9, e109213 (2014).
Kleinstiver, B. P. et al. Genome-wide specificities of CRISPR−Cas Cpf1 nucleases in human cells. Nat. Biotechnol. 34, 869–874 (2016).
Kim, H. K. et al. In vivo high-throughput profiling of CRISPR−Cpf1 activity. Nat. Methods 14, 153–159 (2017).
Jiang, F. & Doudna, J. A. CRISPR–Cas9 structures and mechanisms. Annu. Rev. Biophys. 46, 505–529 (2017).
Yang, W., Lee, J. Y. & Nowotny, M. Making and breaking nucleic acids: two-Mg2+-ion catalysis and substrate specificity. Mol. Cell 22, 5–13 (2006).
Yang, W. An equivalent metal ion in one- and two-metal-ion catalysis. Nat. Struct. Mol. Biol. 15, 1228–1231 (2008).
Yang, W. Nucleases: diversity of structure, function and mechanism. Q. Rev. Biophys. 44, 1–93 (2011).
Steitz, T. A. & Steitz, J. A. A general two-metal-ion mechanism for catalytic RNA. Proc. Natl Acad. Sci. USA 90, 6498–6502 (1993).
Jinek, M. et al. Structures of Cas9 endonucleases reveal RNA-mediated conformational activation. Science 343, 1247997 (2014).
Biertumpfel, C., Yang, W. & Suck, D. Crystal structure of T4 endonuclease VII resolving a Holliday junction. Nature 449, 616–620 (2007).
Li, C. L. et al. DNA binding and cleavage by the periplasmic nuclease Vvn: a novel structure with a known active site. EMBO J. 22, 4014–4025 (2003).
Dagdas, Y. S., Chen, J. S., Sternberg, S. H., Doudna, J. A. & Yildiz, A. A conformational checkpoint between DNA binding and cleavage by CRISPR-Cas9. Sci. Adv. 3 eaao0027 (2017).
Osuka, S. et al. Real-time observation of flexible domain movements in Cas9. Preprint at http://www.biorxiv.org/content/early/2017/03/29/122069 (2017).
Ariyoshi, M. et al. Atomic structure of the RuvC resolvase: a Holliday junction-specific endonuclease from E. coli. Cell 78, 1063–1072 (1994).
Chen, L., Shi, K., Yin, Z. & Aihara, H. Structural asymmetry in the Thermus thermophilus RuvC dimer suggests a basis for sequential strand cleavages during Holliday junction resolution. Nucleic Acids Res. 41, 648–656 (2013).
Gorecka, K. M., Komorowska, W. & Nowotny, M. Crystal structure of RuvC resolvase in complex with Holliday junction substrate. Nucleic Acids Res. 41, 9945–9955 (2013).
Palermo, G., Miao, Y., Walker, R. C., Jinek, M. & McCammon, J. A. Striking plasticity of CRISPR–Cas9 and key role of non-target DNA, as revealed by molecular simulations. ACS Cent. Sci. 2, 756–763 (2016).
Kim, D. et al. Genome-wide analysis reveals specificities of Cpf1 endonucleases in human cells. Nat. Biotechnol. 34, 863–868 (2016).
Ran, F. A. et al. In vivo genome editing using Staphylococcus aureus Cas9. Nature 520, 186–191 (2015).
Chen, J. S. et al. Enhanced proofreading governs CRISPR-Cas9 targeting accuracy. Naturehttp://dx.doi.org/10.1038/nature24268 (2017).
Palermo, G., Miao, Y., Walker, R. C., Jinek, M. & McCammon, J. A. CRISPR-Cas9 conformational activation as elucidated from enhanced molecular simulations. Proc. Natl Acad. Sci. USA 114, 7260–7265 (2017).
Palermo, G. et al. Protospacer adjacent motif-induced allostery activates CRISPR-Cas9. J. Am. Chem. Soc.http://dx.doi.org/10.1021/jacs.7b05313 (2017).
Frock, R. L. et al. Genome-wide detection of DNA double-stranded breaks induced by engineered nucleases. Nat. Biotechnol. 33, 179–186 (2015).
Wang, X. et al. Unbiased detection of off-target cleavage by CRISPR–Cas9 and TALENs using integrase-defective lentiviral vectors. Nat. Biotechnol. 33, 175–178 (2015).
Kim, D. et al. Digenome-seq: genome-wide profiling of CRISPR–Cas9 off-target effects in human cells. Nat. Methods 12, 237–243 (2015).
O’Geen, H., Henry, I. M., Bhakta, M. S., Meckler, J. F. & Segal, D. J. A genome-wide analysis of Cas9 binding specificity using ChIP-seq and targeted sequence capture. Nucleic Acids Res. 43, 3389–3404 (2015).
Horlbeck, M. A. et al. Nucleosomes impede Cas9 access to DNA in vivo and in vitro. eLife 5 e12677 (2016).
Kim, S., Kim, D., Cho, S. W., Kim, J. & Kim, J. S. Highly efficient RNA-guided genome editing in human cells via delivery of purified Cas9 ribonucleoproteins. Genome Res. 24, 1012–1019 (2014).
Lin, S., Staahl, B. T., Alla, R. K. & Doudna, J. A. Enhanced homology-directed human genome engineering by controlled timing of CRISPR–Cas9 delivery. eLife 3, e04766 (2014).
Cho, S. W. et al. Analysis of off-target effects of CRISPR−Cas-derived RNA-guided endonucleases and nickases. Genome Res. 24, 132–141 (2014).
Guilinger, J. P., Thompson, D. B. & Liu, D. R. Fusion of catalytically inactive Cas9 to FokI nuclease improves the specificity of genome modification. Nat. Biotechnol. 32, 577–582 (2014).
Ran, F. A. et al. Double nicking by RNA-guided CRISPR–Cas9 for enhanced genome editing specificity. Cell 154, 1380–1389 (2013).
Mali, P. et al. CAS9 transcriptional activators for target specificity screening and paired nickases for cooperative genome engineering. Nat. Biotechnol. 31, 833–838 (2013).
Tsai, S. Q. et al. Dimeric CRISPR RNA-guided FokI nucleases for highly specific genome editing. Nat. Biotechnol. 32, 569–576 (2014).
Slaymaker, I. M. et al. Rationally engineered Cas9 nucleases with improved specificity. Science 351, 84–88 (2016).
Kleinstiver, B. P. et al. High-fidelity CRISPR–Cas9 nucleases with no detectable genome-wide off-target effects. Nature 529, 490–495 (2016).
Tsai, S. Q. & Joung, J. K. Defining and improving the genome-wide specificities of CRISPR–Cas9 nucleases. Nat. Rev. Genet. 17, 300–312 (2016).
Tycko, J., Myer, V. E. & Hsu, P. D. Methods for optimizing CRISPR–Cas9 genome editing specificity. Mol. Cell 63, 355–370 (2016).
Yee, J. K. Off-target effects of engineered nucleases. FEBS J. 283, 3239–3248 (2016).
Bisaria, N., Jarmoskaite, I. & Herschlag, D. Lessons from enzyme kinetics reveal specificity principles for RNA-guided nucleases in RNA interference and CRISPR-based genome editing. Cell Syst. 4, 21–29 (2017).
Gootenberg, J. S. et al. Nucleic acid detection with CRISPR−Cas13a/C2c2. Science 356, 438–442 (2017).
East-Seletsky, A., O’Connell, M. R., Burstein, D., Knott, G. J. & Doudna, J. A. RNA targeting by functionally orthogonal type VIA CRISPR−Cas enzymes. Mol. Cell 66, 373–383.e3 (2017).
Friedland, A. E. et al. Characterization of Staphylococcus aureus Cas9: a smaller Cas9 for all-in-one adeno-associated virus delivery and paired nickase applications. Genome Biol. 16, 257 (2015).
Kim, E. et al. In vivo genome editing with a small Cas9 orthologue derived from Campylobacter jejuni. Nat. Commun. 8, 14500 (2017).
Hur, J. K. et al. Targeted mutagenesis in mice by electroporation of Cpf1 ribonucleoproteins. Nat. Biotechnol. 34, 807–808 (2016).
Kim, Y. et al. Generation of knockout mice by Cpf1-mediated gene targeting. Nat. Biotechnol. 34, 808–810 (2016).
Tu, M. et al. A ‘new lease of life’: FnCpf1 possesses DNA cleavage activity for genome editing in human cells. Nucleic Acids Res.https://doi.org/10.1093/nar/gkx783 (2017).
Knott, G. J. et al. Guide-bound structures of an RNA-targeting A-cleaving CRISPR-Cas13a enzyme. Nat. Struct. Mol. Biol.https://doi.org/10.1038/nsmb.3466 (2017).
Acknowledgements
The authors thank M. L. Hochstrasser, L. B. Harrington and A. V. Wright for critical reading and valuable input on the manuscript. J.S.C. is a National Science Foundation Graduate Research Fellow and J.A.D. is a Howard Hughes Medical Institute Investigator.
Author information
Authors and Affiliations
Contributions
The authors contributed equally to this manuscript.
Corresponding author
Ethics declarations
Competing interests
J.A.D. is a co-founder of Caribou Biosciences, Editas Medicine and Intellia Therapeutics; a scientific adviser to Caribou Biosciences, Intellia Therapeutics, eFFECTOR Therapeutics and Driver; and executive director of the Innovative Genomics Institute at the University of California Berkeley (UC Berkeley) and the University of California San Francisco (UCSF). J.S.C. and J.A.D. are inventors on UC Berkeley and Howard Hughes Medical Institute patents for clustered regularly interspaced short palindromic repeats (CRISPR) technologies.
Glossary
- Mobile genetic elements
-
DNA sequences that are capable of moving around a genome, including transposons, plasmids and bacteriophages.
- Protospacers
-
Spacer precursors that are captured from foreign DNA and that are complementary to the CRISPR RNA (crRNA) spacer sequence.
- Protospacer adjacent motif sequence
-
(PAM sequence). A short sequence adjacent to the protospacer within foreign DNA. Recognition of the PAM sequence by effector Cas proteins triggers target interference.
- Endonucleolytic cleavage
-
Achieved by Cas proteins that hydrolyse internal phosphodiester bonds within a nucleotide chain.
- Guide RNA
-
An RNA molecule that includes the CRISPR RNA (crRNA) and directs the Cas interference protein to a target site that is complementary to the spacer sequence.
- Interference
-
The final stage of CRISPR immunity that involves RNA-directed cleavage of target nucleic acids by Cas proteins.
- Nuclease
-
An enzyme that catalyses the cleavage of phosphodiester bonds between nucleic acids.
- Helicase
-
An enzyme that unwinds double-stranded DNA using the energy from ATP hydrolysis.
- RuvC nuclease domain
-
Contains an RNase H-like fold and cleaves single-stranded DNA through a two metal mechanism.
- HEPN domain
-
(Higher eukaryotes and prokaryotes nucleotide-binding RNase domain). Contains conserved motifs and functions as an RNase or a non-catalytic RNA-binding domain.
- Ribonuclease
-
An enzyme that hydrolyses the phosphodiester bonds of an RNA backbone.
- Exonuclease
-
An enzyme that hydrolyses phosphodiester bonds, one at a time, from the ends of a nucleic acid chain.
- Single-guide RNA
-
A chimeric RNA molecule in which the CRISPR RNA (crRNA), which contains a sequence complementary to the target DNA, is covalently linked to a trans-activating crRNA (tracrRNA).
- Scissile phosphate
-
The phosphate within the nucleic acid backbone that is cleaved by a nuclease.
- DNA melting
-
The process of DNA strand separation that does not require an external energy source such as ATP.
- RNA strand invasion
-
A process in which the guide RNA segment interrogates a double-stranded DNA (dsDNA) target to initiate DNA unwinding and to form an RNA–DNA heteroduplex.
- R-loop
-
An RNA–DNA heteroduplex and a displaced DNA strand, which is the end result of RNA strand invasion by Cas ribonucleoprotein (RNP) complexes.
- Kinetic inhibition
-
A model in the context of RNA strand invasion that explains how impeding the rate of R-loop formation reduces Cas9 cleavage activity.
- Kinetic Monte Carlo analyses
-
Reveal the time evolution of a given process (that is, stability of the R-loop over time with mismatched substrates or single-guide RNA (sgRNA) variants).
- HNH nuclease domain
-
Contains a histidine–asparagine–histidine (HNH) motif and is the nuclease within Cas9 that hydrolyses the target DNA strand through a one metal ion mechanism.
- Apoprotein
-
The inactive, unbound state of the protein.
- Molecular dynamics simulation
-
(MD simulation). Computer simulations that capture the time evolution of atomic and/or molecular systems by numerically solving Newton's equations of motion.
- Cas9-digested whole-genome sequencing
-
An in vitro method for detecting Cas9 cleavage sites within genomic DNA using whole-genome sequencing.
- cis-cleavage
-
Occurs when the Cas13–CRISPR RNA (crRNA) complex binds a complementary single-stranded RNA (ssRNA) target, which activates the external higher eukaryotes and prokaryotes nucleotide-binding RNase (HEPN) domain catalytic pocket to cleave the bound ssRNA target.
- trans-cleavage
-
Occurs when the Cas13–CRISPR RNA (crRNA) complex binds a complementary single-stranded RNA (ssRNA) target, which activates the external higher eukaryotes and prokaryotes nucleotide-binding RNase (HEPN) domain catalytic pocket to cleave nonspecific ssRNAs in solution.
Rights and permissions
About this article
Cite this article
Chen, J., Doudna, J. The chemistry of Cas9 and its CRISPR colleagues. Nat Rev Chem 1, 0078 (2017). https://doi.org/10.1038/s41570-017-0078
Published:
DOI: https://doi.org/10.1038/s41570-017-0078
This article is cited by
-
Group B Streptococcus Cas9 variants provide insight into programmable gene repression and CRISPR-Cas transcriptional effects
Communications Biology (2023)
-
Programmable Biosensors Based on RNA-Guided CRISPR/Cas Endonuclease
Biological Procedures Online (2022)
-
CRISPR/Cas-Based Modifications for Therapeutic Applications: A Review
Molecular Biotechnology (2022)
-
Inhibition of RNA-binding proteins with small molecules
Nature Reviews Chemistry (2020)
-
Engineering designer beta cells with a CRISPR-Cas9 conjugation platform
Nature Communications (2020)