Abstract
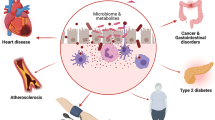
Despite milestones in preventive measures and treatment, cardiovascular disease (CVD) remains associated with a high burden of morbidity and mortality. The protracted nature of the development and progression of CVD motivates the identification of early and complementary targets that might explain and alleviate any residual risk in treated patients. The gut microbiota has emerged as a sentinel between our inner milieu and outer environment and relays a modified risk associated with these factors to the host. Accordingly, numerous mechanistic studies in animal models support a causal role of the gut microbiome in CVD via specific microbial or shared microbiota–host metabolites and have identified converging mammalian targets for these signals. Similarly, large-scale cohort studies have repeatedly reported perturbations of the gut microbial community in CVD, supporting the translational potential of targeting this ecological niche, but the move from bench to bedside has not been smooth. In this Review, we provide an overview of the current evidence on the interconnectedness of the gut microbiome and CVD against the noisy backdrop of highly prevalent confounders in advanced CVD, such as increased metabolic burden and polypharmacy. We further aim to conceptualize the molecular mechanisms at the centre of these associations and identify actionable gut microbiome-based targets, while contextualizing the current knowledge within the clinical scenario and emphasizing the limitations of the field that need to be overcome.
Key points
-
The gut microbiome has been causally linked with cardiometabolic and cardiovascular disorders and is a potential complementary target to understand and reduce the residual risk of cardiovascular disease.
-
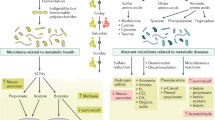
Cardiometabolic and cardiovascular disease progression is reflected in a reduction in gut microbiome diversity and butyrate producers, together with a reduction in pathogen exclusion and increased systemic inflammation.
-
Metabolites produced by gut microbiome metabolism or host–microbiome co-metabolism signal between the gut and peripheral organs to convey a modified exposure and disease risk, providing potential targets for individualized prevention and treatment.
-
A complex interaction exists between medication and the gut microbiome, whereby the microbiome influences drug bioavailability and treatment effects, and medication affects the microbial environment, composition and function, adding both complexity and opportunities for drug repurposing.
-
Next-generation, gut microbiome-based therapeutics require biotic and abiotic characteristics influencing grafting of probiotics, metabolic activity and microbial interactions with the resident bacteria to be addressed; emerging strategies should be paired with improved storage stability, improved viability and targeted delivery.
-
Increasing collaborative efforts to characterize the human gut microbiome diversity in depth are moving us towards the development of more precise and personalized approaches to circumvent disease.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Virani, S. S. et al. Heart disease and stroke statistics — 2020 update: a report from the American Heart Association. Circulation 141, e139–e596 (2020).
Roth, G. A. et al. Global burden of cardiovascular diseases and risk factors, 1990–2019: update from the GBD 2019 study. J. Am. Coll. Cardiol. 76, 2982–3021 (2020).
Fernández-Friera, L. et al. Prevalence, vascular distribution, and multiterritorial extent of subclinical atherosclerosis in a middle-aged cohort. Circulation 131, 2104–2113 (2015).
Ford, E. S., Li, C. & Sattar, N. Metabolic syndrome and incident diabetes: current state of the evidence. Diabetes Care 31, 1898–1904 (2008).
Gami, A. S. et al. Metabolic syndrome and risk of incident cardiovascular events and death: a systematic review and meta-analysis of longitudinal studies. J. Am. Coll. Cardiol. 49, 403–414 (2007).
Koh, K. K., Han, S. H. & Quon, M. J. Inflammatory markers and the metabolic syndrome: insights from therapeutic interventions. J. Am. Coll. Cardiol. 46, 1978–1985 (2005).
Ridker, P. M., Buring, J. E., Cook, N. R. & Rifai, N. C-reactive protein, the metabolic syndrome, and risk of incident cardiovascular events: an 8-year follow-up of 14 719 initially healthy American women. Circulation 107, 391–397 (2003).
Borén, J. et al. Low-density lipoproteins cause atherosclerotic cardiovascular disease: pathophysiological, genetic, and therapeutic insights: a consensus statement from the European Atherosclerosis Society Consensus Panel. Eur. Heart J. 41, 2313–2330 (2020).
Arnett, D. K. et al. 2019 ACC/AHA guideline on the primary prevention of cardiovascular disease: executive summary: a report of the American College of Cardiology/American Heart Association task force on clinical practice guidelines. J. Am. Coll. Cardiol. 74, 1376–1414 (2019).
Sender, R., Fuchs, S. & Milo, R. Are we really vastly outnumbered? Revisiting the ratio of bacterial to host cells in humans. Cell 164, 337–340 (2016).
Rothschild, D. et al. Environment dominates over host genetics in shaping human gut microbiota. Nature 555, 210–215 (2018).
Bäckhed, F. et al. The gut microbiota as an environmental factor that regulates fat storage. Proc. Natl Acad. Sci. USA 101, 15718–15723 (2004).
Ridaura, V. K. et al. Gut microbiota from twins discordant for obesity modulate metabolism in mice. Science 341, 1241214 (2013).
Vrieze, A. et al. Transfer of intestinal microbiota from lean donors increases insulin sensitivity in individuals with metabolic syndrome. Gastroenterology 143, 913–916.e7 (2012).
DeFilipp, Z. et al. Drug-resistant bacteremia transmitted by fecal microbiota transplant. N. Engl. J. Med. 381, 2043–2050 (2019).
Ley, R. E. et al. Obesity alters gut microbial ecology. Proc. Natl Acad. Sci. USA 102, 11070–11075 (2005).
Bäckhed, F., Manchester, J. K., Semenkovich, C. F. & Gordon, J. I. Mechanisms underlying the resistance to diet-induced obesity in germ-free mice. Proc. Natl Acad. Sci. USA 104, 979–984 (2007).
Sze, M. A. & Schloss, P. D. Looking for a signal in the noise: revisiting obesity and the microbiome. mBio 7, e01018-16 (2016).
Walters, W. A., Xu, Z. & Knight, R. Meta-analyses of human gut microbes associated with obesity and IBD. FEBS Lett. 588, 4223–4233 (2014).
Faintuch, J. & Faintuch, S. Microbiome and Metabolome in Diagnosis, Therapy, and other Strategic Applications (Academic, 2019).
Duvallet, C., Gibbons, S. M., Gurry, T., Irizarry, R. A. & Alm, E. J. Meta-analysis of gut microbiome studies identifies disease-specific and shared responses. Nat. Commun. 8, 1784 (2017).
Pasolli, E., Truong, D. T., Malik, F., Waldron, L. & Segata, N. Machine learning meta-analysis of large metagenomic datasets: tools and biological insights. PLoS Comput. Biol. 12, e1004977 (2016).
Duncan, S. H. et al. Reduced dietary intake of carbohydrates by obese subjects results in decreased concentrations of butyrate and butyrate-producing bacteria in feces. Appl. Environ. Microbiol. 73, 1073–1078 (2007).
Waters, J. L. & Ley, R. E. The human gut bacteria Christensenellaceae are widespread, heritable, and associated with health. BMC Biol. 17, 83 (2019).
Goodrich, J. K. et al. Human genetics shape the gut microbiome. Cell 159, 789–799 (2014).
Olsson, L. M. et al. Gut microbiota of obese subjects with Prader-Willi syndrome is linked to metabolic health. Gut 69, 1229–1238 (2020).
Zhong, X. et al. Gut microbiota associations with metabolic health and obesity status in older adults. Nutrients 12, 2364 (2020).
Le Chatelier, E. et al. Richness of human gut microbiome correlates with metabolic markers. Nature 500, 541–546 (2013).
Herder, C. & Roden, M. A novel diabetes typology: towards precision diabetology from pathogenesis to treatment. Diabetologia 65, 1770–1781 (2022).
Zhou, W. et al. Longitudinal multi-omics of host-microbe dynamics in prediabetes. Nature 569, 663–671 (2019).
Wu, H. et al. The gut microbiota in prediabetes and diabetes: a population-based cross-sectional study. Cell Metab. 32, 379–390.e3 (2020).
Schüssler-Fiorenza Rose, S. M. et al. A longitudinal big data approach for precision health. Nat. Med. 25, 792–804 (2019).
Qin, J. et al. A metagenome-wide association study of gut microbiota in type 2 diabetes. Nature 490, 55–60 (2012).
Alvarez-Silva, C. et al. Trans-ethnic gut microbiota signatures of type 2 diabetes in Denmark and India. Genome Med. 13, 37 (2021).
Allin, K. H. et al. Aberrant intestinal microbiota in individuals with prediabetes. Diabetologia 61, 810–820 (2018).
Karlsson, F. H. et al. Gut metagenome in European women with normal, impaired and diabetic glucose control. Nature 498, 99–103 (2013).
Forslund, K. et al. Disentangling type 2 diabetes and metformin treatment signatures in the human gut microbiota. Nature 528, 262–266 (2015).
Forslund, S. K. et al. Combinatorial, additive and dose-dependent drug-microbiome associations. Nature 600, 500–505 (2021).
Olsson, L. M. et al. Dynamics of the normal gut microbiota: a longitudinal one-year population study in Sweden. Cell Host Microbe 30, 726–739.e3 (2022).
Thingholm, L. B. et al. Obese individuals with and without type 2 diabetes show different gut microbial functional capacity and composition. Cell Host Microbe 26, 252–264.e10 (2019).
Belda, E. et al. Impairment of gut microbial biotin metabolism and host biotin status in severe obesity: effect of biotin and prebiotic supplementation on improved metabolism. Gut https://doi.org/10.1136/gutjnl-2021-325753 (2022).
Urpi-Sarda, M. et al. Metabolomics for biomarkers of type 2 diabetes mellitus: advances and nutritional intervention trends. Curr. Cardiovasc. Risk Rep. 9, 280–291 (2015).
Cotillard, A. et al. Dietary intervention impact on gut microbial gene richness. Nature 500, 585–588 (2013).
Fu, J. et al. The gut microbiome contributes to a substantial proportion of the variation in blood lipids. Circ. Res. 117, 817–824 (2015).
Kenny, D. J. et al. Cholesterol metabolism by uncultured human gut bacteria influences host cholesterol level. Cell Host Microbe 28, 245–257.e6 (2020).
Perino, A. & Schoonjans, K. Metabolic messengers: bile acids. Nat. Metab. 4, 416–423 (2022).
Mills, K. T., Stefanescu, A. & He, J. The global epidemiology of hypertension. Nat. Rev. Nephrol. 16, 223–237 (2020).
Jama, H. A., Kaye, D. M. & Marques, F. Z. The gut microbiota and blood pressure in experimental models. Curr. Opin. Nephrol. Hypertens. 28, 97–104 (2019).
Evangelou, E. et al. Genetic analysis of over 1 million people identifies 535 new loci associated with blood pressure traits. Nat. Genet. 50, 1412–1425 (2018).
Verhaar, B. J. H. et al. Associations between gut microbiota, faecal short-chain fatty acids, and blood pressure across ethnic groups: the HELIUS study. Eur. Heart J. 41, 4259–4267 (2020).
Louca, P. et al. Gut microbiome diversity and composition is associated with hypertension in women. J. Hypertens. 39, 1810–1816 (2021).
Sun, S. et al. Gut microbiota composition and blood pressure. Hypertension 73, 998–1006 (2019).
Li, J. et al. Gut microbiota dysbiosis contributes to the development of hypertension. Microbiome 5, 14 (2017).
Palmu, J. et al. Association between the gut microbiota and blood pressure in a population cohort of 6953 individuals. J. Am. Heart Assoc. 9, e016641 (2020).
Menni, C. et al. Gut microbial diversity is associated with lower arterial stiffness in women. Eur. Heart J. 39, 2390–2397 (2018).
Jama, H. A., Beale, A., Shihata, W. A. & Marques, F. Z. The effect of diet on hypertensive pathology: is there a link via gut microbiota-driven immunometabolism? Cardiovasc. Res. 115, 1435–1447 (2019).
Calderón-Pérez, L. et al. Gut metagenomic and short chain fatty acids signature in hypertension: a cross-sectional study. Sci. Rep. 10, 6436 (2020).
Saikku, P. et al. Serological evidence of an association of a novel chlamydia, TWAR, with chronic coronary heart disease and acute myocardial infarction. Lancet 2, 983–986 (1988).
Gelfand, E. V. & Cannon, C. P. Antibiotics for secondary prevention of coronary artery disease: an ACES hypothesis but we need to PROVE IT. Am. Heart J. 147, 202–209 (2004).
Winkel, P. et al. Clarithromycin for stable coronary heart disease increases all-cause and cardiovascular mortality and cerebrovascular morbidity over 10 years in the CLARICOR randomised, blinded clinical trial. Int. J. Cardiol. 182, 459–465 (2015).
Koren, O. et al. Human oral, gut, and plaque microbiota in patients with atherosclerosis. Proc. Natl Acad. Sci. USA 108, 4592–4598 (2011).
Yin, J. et al. Dysbiosis of gut microbiota with reduced trimethylamine-N-oxide level in patients with large-artery atherosclerotic stroke or transient ischemic attack. J. Am. Heart Assoc. 4, e002699 (2015).
Kelly, T. N. et al. Gut microbiome associates with lifetime cardiovascular disease risk profile among Bogalusa Heart Study participants. Circ. Res. 119, 956–964 (2016).
Zhu, Q. et al. Dysbiosis signatures of gut microbiota in coronary artery disease. Physiol. Genomics 50, 893–903 (2018).
Liu, H. et al. Alterations in the gut microbiome and metabolism with coronary artery disease severity. Microbiome 7, 68 (2019).
Toya, T. et al. Coronary artery disease is associated with an altered gut microbiome composition. PLoS ONE 15, e0227147 (2020).
Cui, X. et al. Metagenomic and metabolomic analyses unveil dysbiosis of gut microbiota in chronic heart failure patients. Sci. Rep. 8, 635 (2018).
Karlsson, F. H. et al. Symptomatic atherosclerosis is associated with an altered gut metagenome. Nat. Commun. 3, 1245 (2012).
Feng, Q. et al. Integrated metabolomics and metagenomics analysis of plasma and urine identified microbial metabolites associated with coronary heart disease. Sci. Rep. 6, 22525 (2016).
Jie, Z. et al. The gut microbiome in atherosclerotic cardiovascular disease. Nat. Commun. 8, 845 (2017).
Liu, S., Zhao, W., Liu, X. & Cheng, L. Metagenomic analysis of the gut microbiome in atherosclerosis patients identify cross-cohort microbial signatures and potential therapeutic target. FASEB J. 34, 14166–14181 (2020).
Ott, S. J. et al. Detection of diverse bacterial signatures in atherosclerotic lesions of patients with coronary heart disease. Circulation 113, 929–937 (2006).
Zheng, Y.-Y. et al. Gut microbiome-based diagnostic model to predict coronary artery disease. J. Agric. Food Chem. 68, 3548–3557 (2020).
Talmor-Barkan, Y. et al. Metabolomic and microbiome profiling reveals personalized risk factors for coronary artery disease. Nat. Med. 28, 295–302 (2022).
Fromentin, S. et al. Microbiome and metabolome features of the cardiometabolic disease spectrum. Nat. Med. 28, 303–314 (2022).
Vandeputte, D. et al. Quantitative microbiome profiling links gut community variation to microbial load. Nature 551, 507–511 (2017).
Vieira-Silva, S. et al. Statin therapy is associated with lower prevalence of gut microbiota dysbiosis. Nature 581, 310–315 (2020).
Kasahara, K. et al. Interactions between Roseburia intestinalis and diet modulate atherogenesis in a murine model. Nat. Microbiol. 3, 1461–1471 (2018).
Nemet, I. et al. A cardiovascular disease-linked gut microbial metabolite acts via adrenergic receptors. Cell 180, 862–877.e22 (2020).
Wehedy, E., Shatat, I. F. & Al Khodor, S. The human microbiome in chronic kidney disease: a double-edged sword. Front. Med. 8, 790783 (2021).
Medina, D. A. et al. Cross-regional view of functional and taxonomic microbiota composition in obesity and post-obesity treatment shows country specific microbial contribution. Front. Microbiol. 10, 2346 (2019).
Heianza, Y., Ma, W., Manson, J. E., Rexrode, K. M. & Qi, L. Gut microbiota metabolites and risk of major adverse cardiovascular disease events and death: a systematic review and meta-analysis of prospective studies. J. Am. Heart Assoc. 6, e004947 (2017).
Filippo, C. D. et al. Diet, environments, and gut microbiota. A preliminary investigation in children living in rural and urban Burkina Faso and Italy. Front. Microbiol. 8, 1979 (2017).
Bergman, E. N. Energy contributions of volatile fatty acids from the gastrointestinal tract in various species. Physiol. Rev. 70, 567–590 (1990).
Vinolo, M. A. R., Rodrigues, H. G., Nachbar, R. T. & Curi, R. Regulation of inflammation by short chain fatty acids. Nutrients 3, 858–876 (2011).
Tan, J. et al. The role of short-chain fatty acids in health and disease. Adv. Immunol. 121, 91–119 (2014).
den Besten, G. et al. The role of short-chain fatty acids in the interplay between diet, gut microbiota, and host energy metabolism. J. Lipid Res. 54, 2325–2340 (2013).
Byndloss, M. X. et al. Microbiota-activated PPAR-γ signaling inhibits dysbiotic Enterobacteriaceae expansion. Science 357, 570–575 (2017).
Clemente, J. C. et al. The microbiome of uncontacted Amerindians. Sci. Adv. 1, e1500183 (2015).
Arora, T. & Bäckhed, F. The gut microbiota and metabolic disease: current understanding and future perspectives. J. Intern. Med. 280, 339–349 (2016).
Koh, A., De Vadder, F., Kovatcheva-Datchary, P. & Bäckhed, F. From dietary fiber to host physiology: short-chain fatty acids as key bacterial metabolites. Cell 165, 1332–1345 (2016).
Koh, A. & Bäckhed, F. From association to causality: the role of the gut microbiota and its functional products on host metabolism. Mol. Cell 78, 584–596 (2020).
De Vadder, F. et al. Microbiota-generated metabolites promote metabolic benefits via gut-brain neural circuits. Cell 156, 84–96 (2014).
Mortensen, F. V., Nielsen, H., Mulvany, M. J. & Hessov, I. Short chain fatty acids dilate isolated human colonic resistance arteries. Gut 31, 1391–1394 (1990).
Natarajan, N. et al. Microbial short chain fatty acid metabolites lower blood pressure via endothelial G protein-coupled receptor 41. Physiol. Genomics 48, 826–834 (2016).
Natarajan, N., Hori, D., Berkowitz, D. E. & Pluznick, J. L. Microbial short chain fatty acid (SCFA) metabolites lower blood pressure (BP) via endothelial G-protein coupled receptor 41 (gpr41) [abstract 030]. Hypertension 66, A030 (2015).
Haghikia, A. et al. Propionate attenuates atherosclerosis by immune-dependent regulation of intestinal cholesterol metabolism. Eur. Heart J. 43, 518–533 (2022).
Sonnenburg, E. D. et al. Diet-induced extinctions in the gut microbiota compound over generations. Nature 529, 212–215 (2016).
Boets, E. et al. Systemic availability and metabolism of colonic-derived short-chain fatty acids in healthy subjects: a stable isotope study. J. Physiol. 595, 541–555 (2017).
Senthong, V. et al. Plasma trimethylamine N-oxide, a gut microbe-generated phosphatidylcholine metabolite, is associated with atherosclerotic burden. J. Am. Coll. Cardiol. 67, 2620–2628 (2016).
Tang, W. H. W. et al. Intestinal microbial metabolism of phosphatidylcholine and cardiovascular risk. N. Engl. J. Med. 368, 1575–1584 (2013).
Schiattarella, G. G. et al. Gut microbe-generated metabolite trimethylamine-N-oxide as cardiovascular risk biomarker: a systematic review and dose-response meta-analysis. Eur. Heart J. 38, 2948–2956 (2017).
Haghikia, A. et al. Gut microbiota-dependent trimethylamine N-oxide predicts risk of cardiovascular events in patients with stroke and is related to proinflammatory monocytes. Arterioscler. Thromb. Vasc. Biol. 38, 2225–2235 (2018).
Li, X. S. et al. Gut microbiota-dependent trimethylamine N-oxide in acute coronary syndromes: a prognostic marker for incident cardiovascular events beyond traditional risk factors. Eur. Heart J. 38, 814–824 (2017).
Gregory, J. C. et al. Transmission of atherosclerosis susceptibility with gut microbial transplantation. J. Biol. Chem. 290, 5647–5660 (2015).
Wang, Z. et al. Prognostic value of choline and betaine depends on intestinal microbiota-generated metabolite trimethylamine-N-oxide. Eur. Heart J. 35, 904–910 (2014).
Koeth, R. A. et al. Intestinal microbiota metabolism of L-carnitine, a nutrient in red meat, promotes atherosclerosis. Nat. Med. 19, 576–585 (2013).
Janeiro, M. H., Ramírez, M. J., Milagro, F. I., Martínez, J. A. & Solas, M. Implication of trimethylamine N-oxide (TMAO) in disease: potential biomarker or new therapeutic target. Nutrients 10, 1398 (2018).
Wang, Z. et al. Gut flora metabolism of phosphatidylcholine promotes cardiovascular disease. Nature 472, 57–63 (2011).
Zhu, W. et al. Gut microbial metabolite TMAO enhances platelet hyperreactivity and thrombosis risk. Cell 165, 111–124 (2016).
Ma, G. et al. Trimethylamine N-oxide in atherogenesis: impairing endothelial self-repair capacity and enhancing monocyte adhesion. Biosci. Rep. 37, BSR20160244 (2017).
Chen, K., Zheng, X., Feng, M., Li, D. & Zhang, H. Gut microbiota-dependent metabolite trimethylamine N-oxide contributes to cardiac dysfunction in western diet-induced obese mice. Front. Physiol. 8, 139 (2017).
Farhangi, M. A. & Vajdi, M. Novel findings of the association between gut microbiota-derived metabolite trimethylamine oxide and inflammation: results from a systematic review and dose-response meta-analysis. Crit. Rev. Food Sci. Nutr. 60, 2801–2823 (2020).
Chou, R.-H. et al. Trimethylamine N-oxide, circulating endothelial progenitor cells, and endothelial function in patients with stable angina. Sci. Rep. 9, 4249 (2019).
Tan, Y. et al. Plasma trimethylamine N-oxide as a novel biomarker for plaque rupture in patients with ST-segment-elevation myocardial infarction. Circ. Cardiovasc. Interv. 12, e007281 (2019).
Li, Z. et al. Gut microbe-derived metabolite trimethylamine N-oxide induces cardiac hypertrophy and fibrosis. Lab. Invest. 99, 346–357 (2019).
Jiang, S. et al. Gut microbiota dependent trimethylamine N-oxide aggravates angiotensin II-induced hypertension. Redox Biol. 46, 102115 (2021).
Abbasalizad Farhangi, M. & Vajdi, M. Gut microbiota-associated trimethylamine N-oxide and increased cardiometabolic risk in adults: a systematic review and dose-response meta-analysis. Nutr. Rev. 79, 1022–1042 (2021).
Tang, W. H. W. et al. Gut microbiota-dependent trimethylamine N-oxide (TMAO) pathway contributes to both development of renal insufficiency and mortality risk in chronic kidney disease. Circ. Res. 116, 448–455 (2015).
Papandreou, C., Moré, M. & Bellamine, A. Trimethylamine N-oxide in relation to cardiometabolic health–cause or effect? Nutrients 12, 1330 (2020).
Meyer, K. A. et al. Microbiota-dependent metabolite trimethylamine N-oxide and coronary artery calcium in the coronary artery risk development in young adults study (CARDIA). J. Am. Heart Assoc. 5, e003970 (2016).
Molinaro, A. et al. Imidazole propionate is increased in diabetes and associated with dietary patterns and altered microbial ecology. Nat. Commun. 11, 5881 (2020).
Koh, A. et al. Microbially produced imidazole propionate impairs insulin signaling through mTORC1. Cell 175, 947–961.e17 (2018).
Koh, A. et al. Microbial imidazole propionate affects responses to metformin through p38γ-dependent inhibitory AMPK phosphorylation. Cell Metab. 32, 643–653.e4 (2020).
Liao, Y. et al. Exacerbation of heart failure in adiponectin-deficient mice due to impaired regulation of AMPK and glucose metabolism. Cardiovasc. Res. 67, 705–713 (2005).
Krautkramer, K. A., Fan, J. & Bäckhed, F. Gut microbial metabolites as multi-kingdom intermediates. Nat. Rev. Microbiol. 19, 77–94 (2021).
Molinaro, A., Wahlström, A. & Marschall, H.-U. Role of bile acids in metabolic control. Trends Endocrinol. Metab. 29, 31–41 (2018).
Chong Nguyen, C. et al. Circulating bile acids concentration is predictive of coronary artery disease in human. Sci. Rep. 11, 22661 (2021).
Ryan, P. M., Stanton, C. & Caplice, N. M. Bile acids at the cross-roads of gut microbiome-host cardiometabolic interactions. Diabetol. Metab. Syndr. 9, 102 (2017).
Wu, Q. et al. Suppressing the intestinal farnesoid X receptor/sphingomyelin phosphodiesterase 3 axis decreases atherosclerosis. J. Clin. Invest. 131, e142865 (2021).
Siddiqui, M. S. et al. Impact of obeticholic acid on the lipoprotein profile in patients with non-alcoholic steatohepatitis. J. Hepatol. 72, 25–33 (2020).
Desai, M. S. et al. Bile acid excess induces cardiomyopathy and metabolic dysfunctions in the heart. Hepatology 65, 189–201 (2017).
Fuller, A. T. Is p-aminobenzenesulphonamide the active agent in prontosil therapy? Lancet 229, 194–198 (1937).
Spanogiannopoulos, P., Bess, E. N., Carmody, R. N. & Turnbaugh, P. J. The microbial pharmacists within us: a metagenomic view of xenobiotic metabolism. Nat. Rev. Microbiol. 14, 273–287 (2016).
Klünemann, M. et al. Bioaccumulation of therapeutic drugs by human gut bacteria. Nature 597, 533–538 (2021).
American Diabetes Association Professional Practice Committee. Pharmacologic approaches to glycemic treatment: Standards of Medical Care in Diabetes — 2022. Diabetes Care 45, S125–S143 (2022).
No authors listed. Effect of intensive blood-glucose control with metformin on complications in overweight patients with type 2 diabetes (UKPDS 34). UK Prospective Diabetes Study (UKPDS) Group. Lancet 352, 854–865 (1998).
Buse, J. B. et al. The primary glucose-lowering effect of metformin resides in the gut, not the circulation: results from short-term pharmacokinetic and 12-week dose-ranging studies. Diabetes Care 39, 198–205 (2016).
Wu, H. et al. Metformin alters the gut microbiome of individuals with treatment-naive type 2 diabetes, contributing to the therapeutic effects of the drug. Nat. Med. 23, 850–858 (2017).
Caspary, W. F. et al. Alteration of bile acid metabolism and vitamin-B12-absorption in diabetics on biguanides. Diabetologia 13, 187–193 (1977).
Thomas, C. et al. TGR5-mediated bile acid sensing controls glucose homeostasis. Cell Metab. 10, 167–177 (2009).
Beysen, C. et al. Effect of bile acid sequestrants on glucose metabolism, hepatic de novo lipogenesis, and cholesterol and bile acid kinetics in type 2 diabetes: a randomised controlled study. Diabetologia 55, 432–442 (2012).
Cabreiro, F. et al. Metformin retards aging in C. elegans by altering microbial folate and methionine metabolism. Cell 153, 228–239 (2013).
Reinstatler, L., Qi, Y. P., Williamson, R. S., Garn, J. V. & Oakley, G. P. Jr. Association of biochemical B12 deficiency with metformin therapy and vitamin B12 supplements: the National Health and Nutrition Examination Survey, 1999–2006. Diabetes Care 35, 327–333 (2012).
Wilcock, C. & Bailey, C. J. Accumulation of metformin by tissues of the normal and diabetic mouse. Xenobiotica 24, 49–57 (1994).
Martinez-Guryn, K. et al. Small intestine microbiota regulate host digestive and absorptive adaptive responses to dietary lipids. Cell Host Microbe 23, 458–469.e5 (2018).
Wilmanski, T. et al. Heterogeneity in statin responses explained by variation in the human gut microbiome. Med 3, 388–405.e6 (2022).
Kitamura, S., Sugihara, K., Kuwasako, M. & Tatsumi, K. The role of mammalian intestinal bacteria in the reductive metabolism of zonisamide. J. Pharm. Pharmacol. 49, 253–256 (2011).
Zhao, R. et al. Aspirin reduces colorectal tumor development in mice and gut microbes reduce its bioavailability and chemopreventive effects. Gastroenterology 159, 969–983.e4 (2020).
Kim, I. S. et al. Reduced metabolic activity of gut microbiota by antibiotics can potentiate the antithrombotic effect of aspirin. Biochem. Pharmacol. 122, 72–79 (2016).
Imhann, F. et al. Proton pump inhibitors affect the gut microbiome. Gut 65, 740–748 (2016).
Falony, G. et al. Population-level analysis of gut microbiome variation. Science 352, 560–564 (2016).
Vich Vila, A. et al. Impact of commonly used drugs on the composition and metabolic function of the gut microbiota. Nat. Commun. 11, 362 (2020).
Tsuda, A. et al. Influence of proton-pump inhibitors on the luminal microbiota in the gastrointestinal tract. Clin. Transl. Gastroenterol. 6, e89 (2015).
Maier, L. et al. Extensive impact of non-antibiotic drugs on human gut bacteria. Nature 555, 623–628 (2018).
Cheng, Y.-J. et al. The role of macrolide antibiotics in increasing cardiovascular risk. J. Am. Coll. Cardiol. 66, 2173–2184 (2015).
Gaci, N., Borrel, G., Tottey, W., O’Toole, P. W. & Brugère, J.-F. Archaea and the human gut: new beginning of an old story. World J. Gastroenterol. 20, 16062–16078 (2014).
Haiser, H. J. et al. Predicting and manipulating cardiac drug inactivation by the human gut bacterium Eggerthella lenta. Science 341, 295–298 (2013).
Pryor, R. et al. Host-microbe-drug-nutrient screen identifies bacterial effectors of metformin therapy. Cell 178, 1299–1312.e29 (2019).
Salminen, S. et al. The International Scientific Association of Probiotics and Prebiotics (ISAPP) consensus statement on the definition and scope of postbiotics. Nat. Rev. Gastroenterol. Hepatol. 18, 649–667 (2021).
Makki, K., Deehan, E. C., Walter, J. & Bäckhed, F. The impact of dietary fiber on gut microbiota in host health and disease. Cell Host Microbe 23, 705–715 (2018).
Flint, H. J., Duncan, S. H. & Louis, P. The impact of nutrition on intestinal bacterial communities. Curr. Opin. Microbiol. 38, 59–65 (2017).
Wastyk, H. C. et al. Gut-microbiota-targeted diets modulate human immune status. Cell 184, 4137–4153.e14 (2021).
Fragiadakis, G. K. et al. Long-term dietary intervention reveals resilience of the gut microbiota despite changes in diet and weight. Am. J. Clin. Nutr. 111, 1127–1136 (2020).
Guthrie, L. et al. Impact of a 7-day homogeneous diet on interpersonal variation in human gut microbiomes and metabolomes. Cell Host Microbe 30, 863–874.e4 (2022).
Kolodziejczyk, A. A., Zheng, D. & Elinav, E. Diet-microbiota interactions and personalized nutrition. Nat. Rev. Microbiol. 17, 742–753 (2019).
Desai, M. S. et al. A dietary fiber-deprived gut microbiota degrades the colonic mucus barrier and enhances pathogen susceptibility. Cell 167, 1339–1353.e21 (2016).
Bui, T. P. N. & de Vos, W. M. Next-generation therapeutic bacteria for treatment of obesity, diabetes, and other endocrine diseases. Best Pract. Res. Clin. Endocrinol. Metab. 35, 101504 (2021).
Ochoa-Reparaz, J. & Mangalam, A. K. (eds) The Role of the Gut Microbiota in Health and Inflammatory Diseases (Frontiers, 2020).
Jiang, J. et al. Effects of probiotic supplementation on cardiovascular risk factors in hypercholesterolemia: a systematic review and meta-analysis of randomized clinical trial. J. Funct. Foods 74, 104177 (2020).
Hadi, A., Ghaedi, E., Khalesi, S., Pourmasoumi, M. & Arab, A. Effects of synbiotic consumption on lipid profile: a systematic review and meta-analysis of randomized controlled clinical trials. Eur. J. Nutr. 59, 2857–2874 (2020).
Qu, H., Song, L., Zhang, Y., Gao, Z.-Y. & Shi, D.-Z. The effect of prebiotic products on decreasing adiposity parameters in overweight and obese individuals: a systematic review and meta-analysis. Curr. Med. Chem. 28, 419–431 (2021).
Bock, P. M. et al. The effect of probiotics, prebiotics or synbiotics on metabolic outcomes in individuals with diabetes: a systematic review and meta-analysis. Diabetologia 64, 26–41 (2021).
Lin, M. Y. & Chang, F. J. Antioxidative effect of intestinal bacteria Bifidobacterium longum ATCC 15708 and Lactobacillus acidophilus ATCC 4356. Dig. Dis. Sci. 45, 1617–1622 (2000).
Kim, K.-T. et al. Antioxidant and anti-inflammatory effect and probiotic properties of lactic acid bacteria isolated from canine and feline feces. Microorganisms 9, 1971 (2021).
Malik, M. et al. Lactobacillus plantarum 299v supplementation improves vascular endothelial function and reduces inflammatory biomarkers in men with stable coronary artery disease. Circ. Res. 123, 1091–1102 (2018).
Tolonen, A. C. et al. Synthetic glycans control gut microbiome structure and mitigate colitis in mice. Nat. Commun. 13, 1244 (2022).
Han, N. D. et al. Microbial liberation of N-methylserotonin from orange fiber in gnotobiotic mice and humans. Cell 185, 2495–2509.e11 (2022).
Lawson, C. E. et al. Common principles and best practices for engineering microbiomes. Nat. Rev. Microbiol. 17, 725–741 (2019).
Jimenez, M., Langer, R. & Traverso, G. Microbial therapeutics: new opportunities for drug delivery. J. Exp. Med. 216, 1005–1009 (2019).
Albright, M. B. N. et al. Solutions in microbiome engineering: prioritizing barriers to organism establishment. ISME J. 16, 331–338 (2022).
Feuerstadt, P. et al. SER-109, an oral microbiome therapy for recurrent infection. N. Engl. J. Med. 386, 220–229 (2022).
US National Library of Medicine. ClinicalTrials.gov https://clinicaltrials.gov/ct2/show/NCT03751007 (2021).
Delannoy-Bruno, O. et al. Evaluating microbiome-directed fibre snacks in gnotobiotic mice and humans. Nature 595, 91–95 (2021).
Depommier, C. et al. Supplementation with Akkermansia muciniphila in overweight and obese human volunteers: a proof-of-concept exploratory study. Nat. Med. 25, 1096–1103 (2019).
Arora, T. & Tremaroli, V. Therapeutic potential of butyrate for treatment of type 2 diabetes. Front. Endocrinol. 12, 761834 (2021).
Roshanravan, N. et al. Effect of butyrate and inulin supplementation on glycemic status, lipid profile and glucagon-like peptide 1 level in patients with type 2 diabetes: a randomized double-blind, placebo-controlled trial. Horm. Metab. Res. 49, 886–891 (2017).
Cleophas, M. C. P. et al. Effects of oral butyrate supplementation on inflammatory potential of circulating peripheral blood mononuclear cells in healthy and obese males. Sci. Rep. 9, 775 (2019).
Bouter, K. et al. Differential metabolic effects of oral butyrate treatment in lean versus metabolic syndrome subjects. Clin. Transl. Gastroenterol. 9, 155 (2018).
Yu, Z. et al. Oral supplementation with butyrate improves myocardial ischemia/reperfusion injury via a gut-brain neural circuit. Front. Cardiovasc. Med. 8, 718674 (2021).
Quévrain, E. et al. Identification of an anti-inflammatory protein from Faecalibacterium prausnitzii, a commensal bacterium deficient in Crohn’s disease. Gut 65, 415–425 (2016).
Chambers, E. S. et al. Effects of targeted delivery of propionate to the human colon on appetite regulation, body weight maintenance and adiposity in overweight adults. Gut 64, 1744–1754 (2015).
Pingitore, A. et al. The diet-derived short chain fatty acid propionate improves beta-cell function in humans and stimulates insulin secretion from human islets in vitro. Diabetes Obes. Metab. 19, 257–265 (2017).
Wang, Z. et al. Non-lethal inhibition of gut microbial trimethylamine production for the treatment of atherosclerosis. Cell 163, 1585–1595 (2015).
Roberts, A. B. et al. Development of a gut microbe-targeted nonlethal therapeutic to inhibit thrombosis potential. Nat. Med. 24, 1407–1417 (2018).
Robert, S. et al. Oral delivery of glutamic acid decarboxylase (GAD)-65 and IL10 by Lactococcus lactis reverses diabetes in recent-onset NOD mice. Diabetes 63, 2876–2887 (2014).
Arora, T. et al. Microbially produced glucagon-like peptide 1 improves glucose tolerance in mice. Mol. Metab. 5, 725–730 (2016).
Mimee, M., Tucker, A. C., Voigt, C. A. & Lu, T. K. Programming a human commensal bacterium, Bacteroides thetaiotaomicron, to sense and respond to stimuli in the murine gut microbiota. Cell Syst. 2, 214 (2016).
Bhattarai, Y. et al. Gut microbiota-produced tryptamine activates an epithelial G-protein-coupled receptor to increase colonic secretion. Cell Host Microbe 23, 775–785.e5 (2018).
Bhattarai, Y. et al. Bacterially derived tryptamine increases mucus release by activating a host receptor in a mouse model of inflammatory bowel disease. iScience 23, 101798 (2020).
Funabashi, M. et al. A metabolic pathway for bile acid dehydroxylation by the gut microbiome. Nature 582, 566–570 (2020).
Heianza, Y. et al. Duration and life-stage of antibiotic use and risks of all-cause and cause-specific mortality: prospective cohort study. Circ. Res. 126, 364–373 (2020).
Kwiatek, M., Parasion, S. & Nakonieczna, A. Therapeutic bacteriophages as a rescue treatment for drug-resistant infections – an in vivo studies overview. J. Appl. Microbiol. 128, 985–1002 (2020).
Sutton, T. D. S. & Hill, C. Gut bacteriophage: current understanding and challenges. Front. Endocrinol. 10, 784 (2019).
Lam, K. N. et al. Phage-delivered CRISPR-Cas9 for strain-specific depletion and genomic deletions in the gut microbiome. Cell Rep. 37, 109930 (2021).
Lee, J.-Y., Tsolis, R. M. & Bäumler, A. J. The microbiome and gut homeostasis. Science 377, eabp9960 (2022).
Atarashi, K. et al. Ectopic colonization of oral bacteria in the intestine drives TH1 cell induction and inflammation. Science 358, 359–365 (2017).
Martinez-Guryn, K., Leone, V. & Chang, E. B. Regional diversity of the gastrointestinal microbiome. Cell Host Microbe 26, 314–324 (2019).
Human Microbiome Project Consortium. A framework for human microbiome research. Nature 486, 215–221 (2012).
Chain, P. S. G. et al. Genome Project standards in a new era of sequencing. Science 326, 236–237 (2009).
Costea, P. I. et al. Towards standards for human fecal sample processing in metagenomic studies. Nat. Biotechnol. 35, 1069–1076 (2017).
Sczyrba, A. et al. Critical Assessment of Metagenome Interpretation–a benchmark of metagenomics software. Nat. Methods 14, 1063–1071 (2017).
Wilmanski, T. et al. Blood metabolome predicts gut microbiome α-diversity in humans. Nat. Biotechnol. 37, 1217–1228 (2019).
Pasolli, E. et al. Large-scale genome-wide analysis links lactic acid bacteria from food with the gut microbiome. Nat. Commun. 11, 2610 (2020).
Hildebrand, F. Ultra-resolution metagenomics: when enough is not enough. mSystems 6, e0088121 (2021).
Ahlqvist, E. et al. Novel subgroups of adult-onset diabetes and their association with outcomes: a data-driven cluster analysis of six variables. Lancet Diabetes Endocrinol. 6, 361–369 (2018).
Zou, Y. et al. 1,520 reference genomes from cultivated human gut bacteria enable functional microbiome analyses. Nat. Biotechnol. 37, 179–185 (2019).
Zaneveld, J. R., McMinds, R. & Vega Thurber, R. Stress and stability: applying the Anna Karenina principle to animal microbiomes. Nat. Microbiol. 2, 17121 (2017).
Poyet, M. et al. A library of human gut bacterial isolates paired with longitudinal multiomics data enables mechanistic microbiome research. Nat. Med. 25, 1442–1452 (2019).
Brüssow, H. Problems with the concept of gut microbiota dysbiosis. Microb. Biotechnol. 13, 423–434 (2020).
Debré, P. Louis Pasteur and Claude Bernard: about a posthumous controversy [French]. Biol. Aujourdhui 211, 161–164 (2017).
Gilbert, J. A. & Gibbons, S. M. Integrating microbiomes into clinical trials–the importance of time. BioTechniques https://www.biotechniques.com/microbiology/microbiome_sptl-integrating-microbiomes-into-clinical-trials-the-importance-of-time/ (2020).
Holst, J. J. From the incretin concept and the discovery of GLP-1 to today’s diabetes therapy. Front. Endocrinol. 10, 260 (2019).
Petersen, C. & Round, J. L. Defining dysbiosis and its influence on host immunity and disease. Cell. Microbiol. 16, 1024–1033 (2014).
Parks, D. J. et al. Bile acids: natural ligands for an orphan nuclear receptor. Science 284, 1365–1368 (1999).
Hung, S.-C., Kuo, K.-L., Wu, C.-C. & Tarng, D.-C. Indoxyl sulfate: a novel cardiovascular risk factor in chronic kidney disease. J. Am. Heart Assoc. 6, e005022 (2017).
Tumur, Z. & Niwa, T. Indoxyl sulfate inhibits nitric oxide production and cell viability by inducing oxidative stress in vascular endothelial cells. Am. J. Nephrol. 29, 551–557 (2009).
Tanaka, S. et al. Indoxyl sulfate contributes to adipose tissue inflammation through the activation of NADPH oxidase. Toxins 12, 502 (2020).
Yisireyili, M. et al. Indoxyl sulfate promotes cardiac fibrosis with enhanced oxidative stress in hypertensive rats. Life Sci. 92, 1180–1185 (2013).
Asai, H., Hirata, J. & Watanabe-Akanuma, M. Indoxyl glucuronide, a protein-bound uremic toxin, inhibits hypoxia-inducible factor-dependent erythropoietin expression through activation of aryl hydrocarbon receptor. Biochem. Biophys. Res. Commun. 504, 538–544 (2018).
Karbowska, M. et al. Indoxyl sulfate promotes arterial thrombosis in rat model via increased levels of complex TF/VII, PAI-1, platelet activation as well as decreased contents of SIRT1 and SIRT3. Front. Physiol. 9, 1623 (2018).
Ley, R. E., Lozupone, C. A., Hamady, M., Knight, R. & Gordon, J. I. Worlds within worlds: evolution of the vertebrate gut microbiota. Nat. Rev. Microbiol. 6, 776–788 (2008).
Jankowski, J., Floege, J., Fliser, D., Böhm, M. & Marx, N. Cardiovascular disease in chronic kidney disease: pathophysiological insights and therapeutic options. Circulation 143, 1157–1172 (2021).
Graboski, A. L. & Redinbo, M. R. Gut-derived protein-bound uremic toxins. Toxins 12, 590 (2020).
Natividad, J. M. et al. Impaired aryl hydrocarbon receptor ligand production by the gut microbiota is a key factor in metabolic syndrome. Cell Metab. 28, 737–749.e4 (2018).
Cason, C. A. et al. Plasma microbiome-modulated indole- and phenyl-derived metabolites associate with advanced atherosclerosis and postoperative outcomes. J. Vasc. Surg. 68, 1552–1562.e7 (2018).
Chen, L. et al. The long-term genetic stability and individual specificity of the human gut microbiome. Cell 184, 2302–2315.e12 (2021).
Wu, I.-W. et al. Serum free p-cresyl sulfate levels predict cardiovascular and all-cause mortality in elderly hemodialysis patients – a prospective cohort study. Nephrol. Dial. Transpl. 27, 1169–1175 (2012).
Guerrero, F. et al. Role of endothelial microvesicles released by p-cresol on endothelial dysfunction. Sci. Rep. 10, 10657 (2020).
Han, H. et al. p‐Cresyl sulfate aggravates cardiac dysfunction associated with chronic kidney disease by enhancing apoptosis of cardiomyocytes. J. Am. Heart Assoc. 4, e001852 (2015).
Lin, C.-J., Wu, V., Wu, P.-C. & Wu, C.-J. Meta-analysis of the associations of p-cresyl sulfate (PCS) and indoxyl sulfate (IS) with cardiovascular events and all-cause mortality in patients with chronic renal failure. PLoS ONE 10, e0132589 (2015).
Poesen, R. et al. Microbiota-derived phenylacetylglutamine associates with overall mortality and cardiovascular disease in patients with CKD. J. Am. Soc. Nephrol. 27, 3479–3487 (2016).
Nallu, A., Sharma, S., Ramezani, A., Muralidharan, J. & Raj, D. Gut microbiome in chronic kidney disease: challenges and opportunities. Transl. Res. 179, 24–37 (2017).
Loo, R. L., Zou, X., Appel, L. J., Nicholson, J. K. & Holmes, E. Characterization of metabolic responses to healthy diets and association with blood pressure: application to the optimal macronutrient intake trial for heart health (OmniHeart), a randomized controlled study. Am. J. Clin. Nutr. 107, 323–334 (2018).
Menni, C. et al. Metabolomic study of carotid-femoral pulse-wave velocity in women. J. Hypertens. 33, 791–796 (2015).
Patel, Y. & Joseph, J. Sodium intake and heart failure. Int. J. Mol. Sci. 21, 9474 (2020).
Wilck, N. et al. Salt-responsive gut commensal modulates T17 axis and disease. Nature 551, 585–589 (2017).
Rothhammer, V. et al. Type I interferons and microbial metabolites of tryptophan modulate astrocyte activity and central nervous system inflammation via the aryl hydrocarbon receptor. Nat. Med. 22, 586–597 (2016).
van Nood, E. et al. Duodenal infusion of donor feces for recurrent Clostridium difficile. N. Engl. J. Med. 368, 407–415 (2013).
Moayyedi, P. et al. Fecal microbiota transplantation induces remission in patients with active ulcerative colitis in a randomized controlled trial. Gastroenterology 149, 102–109.e6 (2015).
Costello, S. P. et al. Effect of fecal microbiota transplantation on 8-week remission in patients with ulcerative colitis: a randomized clinical trial. JAMA 321, 156–164 (2019).
Narula, N. et al. Systematic review and meta-analysis: fecal microbiota transplantation for treatment of active ulcerative colitis. Inflamm. Bowel Dis. 23, 1702–1709 (2017).
Rossen, N. G. et al. Findings from a randomized controlled trial of fecal transplantation for patients with ulcerative colitis. Gastroenterology 149, 110–118.e4 (2015).
Kootte, R. S. et al. Improvement of insulin sensitivity after lean donor feces in metabolic syndrome is driven by baseline intestinal microbiota composition. Cell Metab. 26, 611–619.e6 (2017).
Rinott, E. et al. Effects of diet-modulated autologous fecal microbiota transplantation on weight regain. Gastroenterology 160, 158–173.e10 (2021).
Mocanu, V. et al. Fecal microbial transplantation and fiber supplementation in patients with severe obesity and metabolic syndrome: a randomized double-blind, placebo-controlled phase 2 trial. Nat. Med. 27, 1272–1279 (2021).
Smits, L. P. et al. Effect of vegan fecal microbiota transplantation on carnitine- and choline-derived trimethylamine-N-oxide production and vascular inflammation in patients with metabolic syndrome. J. Am. Heart Assoc. 7, e008342 (2018).
Blount, K. F., Shannon, W. D., Deych, E. & Jones, C. Restoration of bacterial microbiome composition and diversity among treatment responders in a phase 2 trial of RBX2660: an investigational microbiome restoration therapeutic. Open Forum Infect. Dis. 6, ofz095 (2019).
Acknowledgements
The authors thank V. Tremaroli and K. Makki (University of Gothenburg, Sweden) for vibrant and rewarding scientific discussions. R.M.C.’s research work is supported by a Walter Benjamin Fellowship grant from the Deutsche Forschungsgemeinschaft (DFG, German Research Foundation). Work in F.B.’s laboratory is supported in part by Transatlantic Networks of Excellence Award from the Leducq Foundation (17CVD01), AFA insurances, Swedish Heart Lung Foundation (20180600), the Knut and Alice Wallenberg Foundation (2017.0026), and grants from the Swedish state under the agreement between the Swedish government and the county councils, the ALF-agreement (ALFGBG- 718101). F.B. is the Torsten Söderberg Professor in Medicine and Wallenberg Scholar.
Author information
Authors and Affiliations
Contributions
R.M.C. researched data for the article, and all the authors contributed to discussion of its content. R.M.C. and F.B. wrote the manuscript, and all the authors reviewed and/or edited it before submission.
Corresponding author
Ethics declarations
Competing interests
F.B. is a shareholder in Implexion Pharma AB, is on the scientific advisory board of Bactolife A/S and receives research funds from BioGaia AB. The other authors declare no competing interests.
Peer review
Peer review information
Nature Reviews Cardiology thanks the anonymous reviewers for their contribution to the peer review of this work.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Related links
Genomic Standards Consortium: https://www.gensc.org/
International Human Microbiome Standards: http://www.human-microbiome.org/index.php
NIH Human Microbiome Project: https://hmpdacc.org/
Glossary
- Microbiota
-
The collection of all living microorganisms present in a specific ecosystem or environment.
- Xenobiotics
-
Any chemical substances found in an organism that are not naturally produced or expected to be present in that organism.
- Microbiome
-
The combination of the microbiota together with elements that are crucial to their function; this ‘theatre of activity’ includes the collective genomes of the microorganisms present.
- Diversity
-
Some high-level measures, such as α-diversity and β-diversity, can be used to describe the microbiome; α-diversity estimates diversity within a single sample, whereas β-diversity describes the diversity (dissimilarity) of two or more communities (samples).
- Richness
-
A measure of α-diversity that refers to the total number of features at a prespecified taxonomic level in a sample or community.
- Opportunistic pathogens
-
Microorganisms that can colonize, but usually do not cause infections, in healthy hosts; under specific conditions, for example in immunocompromised hosts, these microorganisms can cause disease.
- Pathobionts
-
Any potentially pathological organism which, under normal circumstances, lives as a non-harming symbiont; they opportunistically emerge as a result of perturbations in the healthy microbiome owing to complex interactions of genetic, exposomal, microbial and host factors that lead to their selection and expansion.
- Probiotics
-
Live microorganisms selected to provide health benefits when consumed, generally by improving or restoring the gut microbiota.
- Synbiotics
-
Mixtures of probiotics and prebiotics that beneficially affect the host by improving the survival and implantation of live microbial dietary supplements in the gastrointestinal tract.
- Postbiotics
-
Soluble factors (metabolic products or byproducts) that are secreted by live bacteria or are released after bacterial lysis and that provide physiological benefits to the host.
- Obligate anaerobes
-
Bacteria and Archaea that grow only in the absence of oxygen.
Rights and permissions
Springer Nature or its licensor holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Chakaroun, R.M., Olsson, L.M. & Bäckhed, F. The potential of tailoring the gut microbiome to prevent and treat cardiometabolic disease. Nat Rev Cardiol 20, 217–235 (2023). https://doi.org/10.1038/s41569-022-00771-0
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41569-022-00771-0
This article is cited by
-
Cardiovascular disease and cancer: shared risk factors and mechanisms
Nature Reviews Cardiology (2024)
-
Gut bacteria can break down cholesterol
Nature Reviews Cardiology (2024)
-
Ginseng extracts improve circadian clock gene expression and reduce inflammation directly and indirectly through gut microbiota and PI3K signaling pathway
npj Biofilms and Microbiomes (2024)
-
When smoke meets gut: deciphering the interactions between tobacco smoking and gut microbiota in disease development
Science China Life Sciences (2024)
-
Therapeutic applications of gut microbes in cardiometabolic diseases: current state and perspectives
Applied Microbiology and Biotechnology (2024)