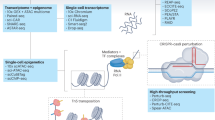
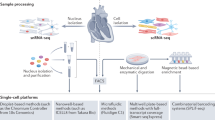
Abstract
Advances in single-cell RNA sequencing (scRNA-seq) technologies in the past 10 years have had a transformative effect on biomedical research, enabling the profiling and analysis of the transcriptomes of single cells at unprecedented resolution and throughput. Specifically, scRNA-seq has facilitated the identification of novel or rare cell types, the analysis of single-cell trajectory construction and stem or progenitor cell differentiation, and the comparison of healthy and disease-related tissues at single-cell resolution. These applications have been critical in advances in cardiovascular research in the past decade as evidenced by the generation of cell atlases of mammalian heart and blood vessels and the elucidation of mechanisms involved in cardiovascular development and stem or progenitor cell differentiation. In this Review, we summarize the currently available scRNA-seq technologies and analytical tools and discuss the latest findings using scRNA-seq that have substantially improved our knowledge on the development of the cardiovascular system and the mechanisms underlying cardiovascular diseases. Furthermore, we examine emerging strategies that integrate multimodal single-cell platforms, focusing on future applications in cardiovascular precision medicine that use single-cell omics approaches to characterize cell-specific responses to drugs or environmental stimuli and to develop effective patient-specific therapeutics.
Key points
-
The advent of single-cell RNA sequencing (scRNA-seq) technologies has facilitated the profiling and analysis of the transcriptomes of single cells at unprecedented resolution and throughput.
-
scRNA-seq allows the identification of rare subpopulations of cells as well as the cellular trajectory analysis of each cell’s transcriptome, helping to identify cell-state transitions during development and progenitor or stem cell differentiation.
-
In addition to the characterization of specific tissues or organ systems, scRNA-seq has also been performed on a larger scale to establish comprehensive cell atlases of various major organs, including the heart.
-
Multimodal single-cell platforms can be integrated and used to evaluate cell population heterogeneity and its contributions to patient-specific drug responses and adverse effects.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Heid, C. A., Stevens, J., Livak, K. J. & Williams, P. M. Real time quantitative PCR. Genome Res. 6, 986–994 (1996).
Trapnell, C. Defining cell types and states with single-cell genomics. Genome Res. 25, 1491–1498 (2015).
Islam, S. et al. Quantitative single-cell RNA-seq with unique molecular identifiers. Nat. Methods 11, 163–166 (2014).
Shapiro, E., Biezuner, T. & Linnarsson, S. Single-cell sequencing-based technologies will revolutionize whole-organism science. Nat. Rev. Genet. 14, 618–630 (2013).
Potter, S. S. Single-cell RNA sequencing for the study of development, physiology and disease. Nat. Rev. Nephrol. 14, 479–492 (2018).
The Tabula Muris Consortium et al. Single-cell transcriptomics of 20 mouse organs creates a Tabula Muris. Nature 562, 367–372 (2018).
Han, X. et al. Mapping the Mouse Cell Atlas by Microwell-Seq. Cell 172, 1091–1107.e17 (2018).
Cusanovich, D. A. et al. A single-cell atlas of in vivo mammalian chromatin accessibility. Cell 174, 1309–1324.e18 (2018).
Gao, S. et al. Tracing the temporal-spatial transcriptome landscapes of the human fetal digestive tract using single-cell RNA-sequencing. Nat. Cell Biol. 20, 721–734 (2018).
Wang, P. et al. Dissecting the global dynamic molecular profiles of human fetal kidney development by single-cell RNA sequencing. Cell Rep. 24, 3554–3567.e3 (2018).
Zhong, S. et al. A single-cell RNA-seq survey of the developmental landscape of the human prefrontal cortex. Nature 555, 524–528 (2018).
Cui, Y. et al. Single-cell transcriptome analysis maps the developmental track of the human heart. Cell Rep. 26, 1934–1950.e5 (2019).
Ramilowski, J. A. et al. A draft network of ligand-receptor-mediated multicellular signalling in human. Nat. Commun. 6, 7866 (2015).
Ståhl, P. L. et al. Visualization and analysis of gene expression in tissue sections by spatial transcriptomics. Science 353, 78–82 (2016).
Jemt, A. et al. An automated approach to prepare tissue-derived spatially barcoded RNA-sequencing libraries. Sci. Rep. 6, 37137 (2016).
Asp, M. et al. Spatial detection of fetal marker genes expressed at low level in adult human heart tissue. Sci. Rep. 7, 12941 (2017).
Berglund, E. et al. Spatial maps of prostate cancer transcriptomes reveal an unexplored landscape of heterogeneity. Nat. Commun. 9, 2419 (2018).
Asp, M. et al. A spatiotemporal organ-wide gene expression and cell atlas of the developing human heart. Cell 179, 1647–1660.e19 (2019).
Svensson, V., Vento-Tormo, R. & Teichmann, S. A. Exponential scaling of single-cell RNA-seq in the past decade. Nat. Protoc. 13, 599–604 (2018).
Macosko, E. Z. et al. Highly parallel genome-wide expression profiling of individual cells using nanoliter droplets. Cell 161, 1202–1214 (2015).
Gladka, M. M. et al. Single-cell sequencing of the healthy and diseased heart reveals cytoskeleton-associated protein 4 as a new modulator of fibroblasts activation. Circulation 138, 166–180 (2018).
Ramsköld, D. et al. Full-length mRNA-Seq from single-cell levels of RNA and individual circulating tumor cells. Nat. Biotechnol. 30, 777–782 (2012).
Picelli, S. et al. Full-length RNA-seq from single cells using Smart-seq2. Nat. Protoc. 9, 171–181 (2014).
Picelli, S. Full-length single-cell RNA sequencing with Smart-seq2. Methods Mol. Biol. 1979, 25–44 (2019).
Hwang, B., Lee, J. H. & Bang, D. Single-cell RNA sequencing technologies and bioinformatics pipelines. Exp. Mol. Med. 50, 96 (2018).
Hashimshony, T., Wagner, F., Sher, N. & Yanai, I. CEL-seq: single-cell RNA-seq by multiplexed linear amplification. Cell Rep. 2, 666–673 (2012).
Hashimshony, T. et al. CEL-seq2: sensitive highly-multiplexed single-cell RNA-seq. Genome Biol. 17, 77 (2016).
Zheng, G. X. Y. et al. Massively parallel digital transcriptional profiling of single cells. Nat. Commun. 8, 14049 (2017).
Papalexi, E. & Satija, R. Single-cell RNA sequencing to explore immune cell heterogeneity. Nat. Rev. Immunol. 18, 35–45 (2018).
Zappia, L., Phipson, B. & Oshlack, A. Exploring the single-cell RNA-seq analysis landscape with the scRNA-tools database. PLoS Comput. Biol. 14, e1006245 (2018).
Nguyen, Q. H., Pervolarakis, N., Nee, K. & Kessenbrock, K. Experimental considerations for single-cell RNA sequencing approaches. Front. Cell Dev. Biol. 6, 108 (2018).
Vieira Braga, F. A. & Miragaia, R. J. in Single Cell Methods: Sequencing and Proteomics (ed. Proserpio, V.) 9–21 (Springer, 2019).
See, K. et al. Single cardiomyocyte nuclear transcriptomes reveal a lincRNA-regulated de-differentiation and cell cycle stress-response in vivo. Nat. Commun. 8, 225 (2017).
Hu, P. et al. Single-nucleus transcriptomic survey of cell diversity and functional maturation in postnatal mammalian hearts. Genes. Dev. 32, 1344–1357 (2018).
Linscheid, N. et al. Quantitative proteomics and single-nucleus transcriptomics of the sinus node elucidates the foundation of cardiac pacemaking. Nat. Commun. 10, 2889 (2019).
Kannan, S. et al. Large particle fluorescence-activated cell sorting enables high-quality single-cell RNA sequencing and functional analysis of adult cardiomyocytes. Circ. Res. 125, 567–569 (2019).
DeLuca, D. S. et al. RNA-SeQC: RNA-seq metrics for quality control and process optimization. Bioinformatics 28, 1530–1532 (2012).
Bacher, R. & Kendziorski, C. Design and computational analysis of single-cell RNA-sequencing experiments. Genome Biol. 17, 63 (2016).
Trapnell, C. et al. The dynamics and regulators of cell fate decisions are revealed by pseudotemporal ordering of single cells. Nat. Biotechnol. 32, 381–386 (2014).
Wolf, F. A., Angerer, P. & Theis, F. J. SCANPY: large-scale single-cell gene expression data analysis. Genome Biol. 19, 15 (2018).
McCarthy, D. J., Campbell, K. R., Lun, A. T. L. & Wills, Q. F. Scater: pre-processing, quality control, normalization and visualization of single-cell RNA-seq data in R. Bioinformatics 33, 1179–1186 (2017).
Diaz, A. et al. SCell: integrated analysis of single-cell RNA-seq data. Bioinformatics 32, 2219–2220 (2016).
Butler, A., Hoffman, P., Smibert, P., Papalexi, E. & Satija, R. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat. Biotechnol. 36, 411–420 (2018).
Guo, M., Wang, H., Potter, S. S., Whitsett, J. A. & Xu, Y. SINCERA: a pipeline for single-cell RNA-seq profiling analysis. PLoS Comput. Biol. 11, e1004575 (2015).
Stegle, O., Teichmann, S. A. & Marioni, J. C. Computational and analytical challenges in single-cell transcriptomics. Nat. Rev. Genet. 16, 133–145 (2015).
Kiselev, V. Y., Andrews, T. S. & Hemberg, M. Challenges in unsupervised clustering of single-cell RNA-seq data. Nat. Rev. Genet. 20, 273–282 (2019).
van der Maaten, L. J. P. & Hinton, G. E. Visualizing high-dimensional data using t-SNE. J. Mach. Learn. Res. 9, 2579–2605 (2008).
Becht, E. et al. Dimensionality reduction for visualizing single-cell data using UMAP. Nat. Biotechnol. 37, 38–44 (2019).
Crow, M., Paul, A., Ballouz, S., Huang, Z. J. & Gillis, J. Characterizing the replicability of cell types defined by single cell RNA-sequencing data using MetaNeighbor. Nat. Commun. 9, 884 (2018).
Andrews, T. S. & Hemberg, M. Identifying cell populations with scRNASeq. Mol. Asp. Med. 59, 114–122 (2018).
Lin, P., Troup, M. & Ho, J. W. K. CIDR: Ultrafast and accurate clustering through imputation for single-cell RNA-seq data. Genome Biol. 18, 59 (2017).
Žurauskienė, J. & Yau, C. pcaReduce: hierarchical clustering of single cell transcriptional profiles. BMC Bioinformatics 17, 140 (2016).
Levine, J. H. et al. Data-driven phenotypic dissection of AML reveals progenitor-like cells that correlate with prognosis. Cell 162, 184–197 (2015).
Raykov, Y. P., Boukouvalas, A., Baig, F. & Little, M. A. What to do when K-means clustering fails: a simple yet principled alternative algorithm. PLoS One 11, e0162259 (2016).
Tanay, A. & Regev, A. Scaling single-cell genomics from phenomenology to mechanism. Nature 541, 331–338 (2017).
Kester, L. & van Oudenaarden, A. Single-cell transcriptomics meets lineage tracing. Cell Stem Cell 23, 166–179 (2018).
Su, T. et al. Single-cell analysis of early progenitor cells that build coronary arteries. Nature 559, 356–362 (2018).
Wang, W. et al. A single-cell transcriptional roadmap for cardiopharyngeal fate diversification. Nat. Cell Biol. 21, 674–686 (2019).
Ludwig, L. S. et al. Lineage tracing in humans enabled by mitochondrial mutations and single-cell genomics. Cell 176, 1325–1339.e22 (2019).
Xu, J. et al. Single-cell lineage tracing by endogenous mutations enriched in transposase accessible mitochondrial DNA. eLife 8, e45105 (2019).
Skelly, D. A. et al. Single-cell transcriptional profiling reveals cellular diversity and intercommunication in the mouse heart. Cell Rep. 22, 600–610 (2018).
Li, G. et al. Single cell expression analysis reveals anatomical and cell cycle-dependent transcriptional shifts during heart development. Development https://doi.org/10.1242/dev.173476 (2019).
Paik, D. T. et al. Large-scale single-cell RNA-seq reveals molecular signatures of heterogeneous populations of human induced pluripotent stem cell-derived endothelial cells. Circ. Res. 123, 443–450 (2018).
Li, G. et al. Transcriptomic profiling maps anatomically patterned subpopulations among single embryonic cardiac cells. Dev. Cell 39, 491–507 (2016).
DeLaughter, D. M. et al. Single-cell resolution of temporal gene expression during heart development. Dev. Cell 39, 480–490 (2016).
Lescroart, F. et al. Defining the earliest step of cardiovascular lineage segregation by single-cell RNA-seq. Science 359, 1177–1181 (2018).
Devine, W. P., Wythe, J. D., George, M., Koshiba-Takeuchi, K. & Bruneau, B. G. Early patterning and specification of cardiac progenitors in gastrulating mesoderm. eLife 3, e03848 (2014).
Jia, G. et al. Single cell RNA-seq and ATAC-seq analysis of cardiac progenitor cell transition states and lineage settlement. Nat. Commun. 9, 4877 (2018).
Xiong, H. et al. Single-cell transcriptomics reveals chemotaxis-mediated intraorgan crosstalk during cardiogenesis. Circ. Res. 125, 398–410 (2019).
de Soysa, T. Y. et al. Single-cell analysis of cardiogenesis reveals basis for organ-level developmental defects. Nature 572, 120–124 (2019).
Muraro, M. J. et al. A single-cell transcriptome atlas of the human pancreas. Cell Syst. 3, 385–394.e3 (2016).
Ackers-Johnson, M., Tan, W. L. W. & Foo, R. S.-Y. Following hearts, one cell at a time: recent applications of single-cell RNA sequencing to the understanding of heart disease. Nat. Commun. 9, 4434 (2018).
Coppini, R. et al. Isolation and functional characterization of human ventricular cardiomyocytes from fresh surgical samples. J. Vis. Exp. https://doi.org/10.3791/51116 (2014).
Nomura, S. et al. Cardiomyocyte gene programs encoding morphological and functional signatures in cardiac hypertrophy and failure. Nat. Commun. 9, 4435 (2018).
Honkoop, H. et al. Single-cell analysis uncovers that metabolic reprogramming by ErbB2 signaling is essential for cardiomyocyte proliferation in the regenerating heart. eLife 8, e50163 (2019).
Li, Z. et al. Single-cell transcriptome analyses reveal novel targets modulating cardiac neovascularization by resident endothelial cells following myocardial infarction. Eur. Heart J. 40, 2507–2520 (2019).
Paik, D. T. et al. Wnt10b gain-of-function improves cardiac repair by arteriole formation and attenuation of fibrosis. Circ. Res. 117, 804–816 (2015).
Farbehi, N. et al. Single-cell expression profiling reveals dynamic flux of cardiac stromal, vascular and immune cells in health and injury. eLife 8, e43882 (2019).
Zeisberg, E. M. et al. Endothelial-to-mesenchymal transition contributes to cardiac fibrosis. Nat. Med. 13, 952–961 (2007).
Aisagbonhi, O. et al. Experimental myocardial infarction triggers canonical Wnt signaling and endothelial-to-mesenchymal transition. Dis. Model. Mech. 4, 469–483 (2011).
Ubil, E. et al. Mesenchymal-endothelial transition contributes to cardiac neovascularization. Nature 514, 585–590 (2014).
Tucker, N. R. et al. Transcriptional and cellular diversity of the human heart. Preprint at bioRxiv https://doi.org/10.1101/2020.01.06.896076 (2020).
Red-Horse, K., Ueno, H., Weissman, I. L. & Krasnow, M. Coronary arteries form by developmental reprogramming of venous cells. Nature 464, 549–553 (2010).
Sharma, B., Chang, A. & Red-Horse, K. Coronary artery development: progenitor cells and differentiation pathways. Annu. Rev. Physiol. 79, 1–19 (2017).
Kalluri, A. S. et al. Single cell analysis of the normal mouse aorta reveals functionally distinct endothelial cell populations. Circulation 140, 147–163 (2019).
McDonald, A. I. et al. Endothelial regeneration of large vessels is a biphasic process driven by local cells with distinct proliferative capacities. Cell Stem Cell 23, 210–225.e6 (2018).
Kuznetsova, T., Prange, K. H. M., Glass, C. K. & de Winther, M. P. J. Transcriptional and epigenetic regulation of macrophages in atherosclerosis. Nat. Rev. Cardiol. https://doi.org/10.1038/s41569-019-0265-3 (2019).
Souilhol, C. et al. Endothelial responses to shear stress in atherosclerosis: a novel role for developmental genes. Nat. Rev. Cardiol. 17, 52–63 (2020).
Basatemur, G. L., Jørgensen, H. F., Clarke, M. C. H., Bennett, M. R. & Mallat, Z. Vascular smooth muscle cells in atherosclerosis. Nat. Rev. Cardiol. 16, 727–744 (2019).
Libby, P. et al. Atherosclerosis. Nat. Rev. Dis. Prim. 5, 56 (2019).
Wirka, R. C. et al. Atheroprotective roles of smooth muscle cell phenotypic modulation and the TCF21 disease gene as revealed by single-cell analysis. Nat. Med. 25, 1280–1289 (2019).
Yao, F. et al. Histone variant H2A.Z is required for the maintenance of smooth muscle cell identity as revealed by single-cell transcriptomics. Circulation 138, 2274–2288 (2018).
Cochain, C. et al. Single-cell RNA-seq reveals the transcriptional landscape and heterogeneity of aortic macrophages in murine atherosclerosis. Circ. Res. 122, 1661–1674 (2018).
Winkels, H. et al. Atlas of the immune cell repertoire in mouse atherosclerosis defined by single-cell RNA-sequencing and mass cytometry. Circ. Res. 122, 1675–1688 (2018).
Lukowski, S. W. et al. Single-cell transcriptional profiling of aortic endothelium identifies a hierarchy from endovascular progenitors to differentiated cells. Cell Rep. 27, 2748–2758.e3 (2019).
Cao, J. et al. The single-cell transcriptional landscape of mammalian organogenesis. Nature 566, 496–502 (2019).
Regev, A. et al. The Human Cell Atlas. eLife 6, e27041 (2017).
HuBMAP Consortium. The human body at cellular resolution: the NIH human biomolecular atlas program. Nature 574, 187–192 (2019).
Franzén, O., Gan, L.-M. & Björkegren, J. L. M. PanglaoDB: a web server for exploration of mouse and human single-cell RNA sequencing data. Database 2019, baz046 (2019).
Shi, Y., Inoue, H., Wu, J. C. & Yamanaka, S. Induced pluripotent stem cell technology: a decade of progress. Nat. Rev. Drug. Discov. 16, 115–130 (2017).
Paik, D. T., Chandy, M. & Wu, J. C. Patient and disease-specific induced pluripotent stem cells for discovery of personalized cardiovascular drugs and therapeutics. Pharmacol. Rev. 72, 320–342 (2020).
Bedada, F. B., Wheelwright, M. & Metzger, J. M. Maturation status of sarcomere structure and function in human iPSC-derived cardiac myocytes. Biochim. Biophys. Acta 1863, 1829–1838 (2016).
Friedman, C. E. et al. Single-cell transcriptomic analysis of cardiac differentiation from human PSCs reveals HOPX-dependent cardiomyocyte maturation. Cell Stem Cell 23, 586–598.e8 (2018).
Churko, J. M. et al. Defining human cardiac transcription factor hierarchies using integrated single-cell heterogeneity analysis. Nat. Commun. 9, 4906 (2018).
Gu, M. et al. Patient-specific iPSC-derived endothelial cells uncover pathways that protect against pulmonary hypertension in BMPR2 mutation carriers. Cell Stem Cell 20, 490–504.e5 (2017).
McCracken, I. R. et al. Transcriptional dynamics of pluripotent stem cell-derived endothelial cell differentiation revealed by single-cell RNA sequencing. Eur. Heart J. 41, 1024–1036 (2020).
Williams, I. M. & Wu, J. C. Generation of endothelial cells from human pluripotent stem cells. Arterioscler. Thromb. Vasc. Biol. 39, 1317–1329 (2019).
Cao, J. et al. Comprehensive single-cell transcriptional profiling of a multicellular organism. Science 357, 661–667 (2017).
Rosenberg, A. B. et al. Single-cell profiling of the developing mouse brain and spinal cord with split-pool barcoding. Science 360, 176–182 (2018).
Efremova, M. & Teichmann, S. A. Computational methods for single-cell omics across modalities. Nat. Methods 17, 14–17 (2020).
Buenrostro, J. D. et al. Single-cell chromatin accessibility reveals principles of regulatory variation. Nature 523, 486–490 (2015).
Satpathy, A. T. et al. Massively parallel single-cell chromatin landscapes of human immune cell development and intratumoral T cell exhaustion. Nat. Biotechnol. 37, 925–936 (2019).
Lareau, C. A. et al. Droplet-based combinatorial indexing for massive-scale single-cell chromatin accessibility. Nat. Biotechnol. 37, 916–924 (2019).
Cusanovich, D. A. et al. The cis-regulatory dynamics of embryonic development at single-cell resolution. Nature 555, 538–542 (2018).
Corces, M. R. et al. Lineage-specific and single-cell chromatin accessibility charts human hematopoiesis and leukemia evolution. Nat. Genet. 48, 1193–1203 (2016).
Satpathy, A. T. et al. Transcript-indexed ATAC-seq for precision immune profiling. Nat. Med. 24, 580–590 (2018).
Cho, S., Irianto, J. & Discher, D. E. Mechanosensing by the nucleus: from pathways to scaling relationships. J. Cell Biol. 216, 305–315 (2017).
Cho, S. et al. Mechanosensing by the lamina protects against nuclear rupture, DNA damage, and cell-cycle arrest. Dev. Cell 49, 920–935.e5 (2019).
Cao, J. et al. Joint profiling of chromatin accessibility and gene expression in thousands of single cells. Science 361, 1380–1385 (2018).
Karemaker, I. D. & Vermeulen, M. Single-cell DNA methylation profiling: technologies and biological applications. Trends Biotechnol. 36, 952–965 (2018).
Angermueller, C. et al. Parallel single-cell sequencing links transcriptional and epigenetic heterogeneity. Nat. Methods 13, 229–232 (2016).
Cheow, L. F. et al. Single-cell multimodal profiling reveals cellular epigenetic heterogeneity. Nat. Methods 13, 833–836 (2016).
Pott, S. Simultaneous measurement of chromatin accessibility, DNA methylation, and nucleosome phasing in single cells. eLife 6, 23203 (2017).
Clark, S. J. et al. scNMT-seq enables joint profiling of chromatin accessibility DNA methylation and transcription in single cells. Nat. Commun. 9, 781 (2018).
Stoeckius, M. et al. Simultaneous epitope and transcriptome measurement in single cells. Nat. Methods. 14, 865–868 (2017).
Specht, H. et al. Single-cell mass-spectrometry quantifies the emergence of macrophage heterogeneity. Preprint at bioRxiv https://doi.org/10.1101/665307 (2019).
Swaminathan, J. et al. Highly parallel single-molecule identification of proteins in zeptomole-scale mixtures. Nat. Biotechnol. 36, 1076–1082 (2018).
Salmén, F. et al. Barcoded solid-phase RNA capture for spatial transcriptomics profiling in mammalian tissue sections. Nat. Protoc. 13, 2501–2534 (2018).
Rodriques, S. G. et al. Slide-seq: a scalable technology for measuring genome-wide expression at high spatial resolution. Science 363, 1463–1467 (2019).
Shalek, A. K. & Benson, M. Single-cell analyses to tailor treatments. Sci. Transl Med. 9, aan4730 (2017).
Levitin, H. M., Yuan, J. & Sims, P. A. Single-cell transcriptomic analysis of tumor heterogeneity. Trends Cancer 4, 264–268 (2018).
Kim, K.-T. et al. Application of single-cell RNA sequencing in optimizing a combinatorial therapeutic strategy in metastatic renal cell carcinoma. Genome Biol. 17, 80 (2016).
Tirosh, I. et al. Dissecting the multicellular ecosystem of metastatic melanoma by single-cell RNA-seq. Science 352, 189–196 (2016).
Lavin, Y. et al. Innate immune landscape in early lung adenocarcinoma by paired single-cell analyses. Cell 169, 750–765.e17 (2017).
Kolandaivelu, K., Leiden, B. B., O’Gara, P. T. & Bhatt, D. L. Non-adherence to cardiovascular medications. Eur. Heart J. 35, 3267–3276 (2014).
Matsa, E. et al. Transcriptome profiling of patient-specific human iPSC-cardiomyocytes predicts individual drug safety and efficacy responses in vitro. Cell Stem Cell 19, 311–325 (2016).
Lam, C. K. et al. Identifying the transcriptome signatures of calcium channel blockers in human induced pluripotent stem cell-derived cardiomyocytes. Circ. Res. 125, 212–222 (2019).
Strzelecka, P. M., Ranzoni, A. M. & Cvejic, A. Dissecting human disease with single-cell omics: application in model systems and in the clinic. Dis. Models Mech. 11, 036525 (2018).
Stuart, T. et al. Comprehensive integration of single-cell data. Cell 177, 1888–1902.e21 (2019).
Jaitin, D. A. et al. Massively parallel single-cell RNA-seq for marker-free decomposition of tissues into cell types. Science 343, 776–779 (2014).
Bagnoli, J. W. et al. Sensitive and powerful single-cell RNA sequencing using mcSCRB-seq. Nat. Commun. 9, 2937 (2018).
Goldstein, L. D. et al. Massively parallel nanowell-based single-cell gene expression profiling. BMC Genomics 18, 519 (2017).
Goodyer, W. R. et al. Transcriptomic profiling of the developing cardiac conduction system at single-cell resolution. Circ. Res. 125, 379–397 (2019).
Tang, J. et al. Arterial Sca1+ vascular stem cells generate de novo smooth muscle for artery repair and regeneration. Cell Stem Cell 26, 81–96.e4 (2020).
Martini, E. et al. Single-cell sequencing of mouse heart immune infiltrate in pressure overload-driven heart failure reveals extent of immune activation. Circulation 140, 2089–2107 (2019).
Francesconi, M. et al. Single cell RNA-seq identifies the origins of heterogeneity in efficient cell transdifferentiation and reprogramming. eLife 8, 41627 (2019).
Nguyen, Q. H. et al. Single-cell RNA-seq of human induced pluripotent stem cells reveals cellular heterogeneity and cell state transitions between subpopulations. Genome Res. 28, 1053–1066 (2018).
Ohta, R. et al. Laminin-guided highly efficient endothelial commitment from human pluripotent stem cells. Sci. Rep. 6, 35680 (2016).
Acknowledgements
The authors are supported by grants from the NIH: K99 HL150216 (D.T.P.), RM1 HG007735 (H.Y.C.), R01 HL130020 (J.C.W.), R01 HL145676 (J.C.W.) and P01 HL141084 (J.C.W.), and from the Leducq Foundation: 18CVD05 (J.C.W.). The authors thank B. C. Wu (Stanford University, USA), J. X. Zhang (Stanford University, USA) and H. Lee (Stanford University, USA) for critical reading of the manuscript.
Author information
Authors and Affiliations
Contributions
D.T.P. and L.T. researched data for the article, D.T.P., H.Y.C. and J.C.W. substantially contributed to the discussion of its content, and D.T.P., S.C. and L.T. wrote, reviewed and edited the manuscript before submission.
Corresponding authors
Ethics declarations
Competing interests
H.Y.C. is a co-founder of Accent Therapeutics and Boundless Bio, and an adviser to 10x Genomics, Arsenal Biosciences and Spring Discovery. J.C.W. is a co-founder of Khloris Biosciences, but has no competing interests, as the work presented here is completely independent. The other authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Related links
Human Cell Atlas: https://www.humancellatlas.org/
PanglaoDB: https://panglaodb.se/
scRNASeqDB: https://bioinfo.uth.edu/scrnaseqdb/
Single Cell Portal: https://singlecell.broadinstitute.org/single_cell
Tabula Muris: https://tabula-muris.ds.czbiohub.org/
The Human BioMolecular Atlas Program: https://commonfund.nih.gov/hubmap
Glossary
- Quantitative PCR
-
A polymerase chain reaction (PCR) that records product expression in real time.
- Microarray
-
A chip containing thousands of wells with a bound DNA of known sequence, which can be used to bind and measure the expression of transcriptome mRNA.
- Bulk RNA sequencing
-
Bulk resolution, next-generation sequencing, which reveals RNA presence and quantity in a sample of cells during time of measurement.
- Transcriptome
-
All RNA molecules expressed in a cell or cell population.
- Cellular trajectory analysis
-
Computational analysis technique used to track and group cells based on their course through a dynamic process such as cell differentiation or the cell cycle.
- Fluorescence-activated cell sorting
-
(FACS). Technique in which target cell types in suspension are separated and sorted by flow cytometry based on fluorescence information.
- R packages
-
Single-cell gene-expression analysis software package written in R that can be run in integrated development environments, such as RStudio.
- Python packages
-
Single-cell gene-expression analysis software package written in Python that can be run in integrated development environments such as Sublime Text or Visual Studio.
- Principal component analysis
-
(PCA). A linear statistical technique that reduces the number of experimental variables to the minimum amount.
- t-Distributed stochastic neighbour embedding
-
(tSNA). Nonlinear variable reduction method that displays high-dimension data points, such as cell transcriptome data, on 2D or 3D graphs, primarily separating points on the basis of (dis)similarity to each other.
- Uniform manifold approximation and projection
-
(UMAP). Nonlinear variable reduction method that displays high-dimension data points on 2D or 3D distance-dependent graphs, which can be used to reveal information such as cell differentiation trajectory and cell state.
- Euclidean distance
-
A measurement of difference or dissimilarity between a pair of samples in an n-dimensional feature space.
- Induced pluripotent stem cells
-
(iPSCs). Pluripotent stem cells reprogrammed from adult somatic cells.
- Immunohistochemistry
-
Antibody-based detection method of protein in samples of tissue.
- In situ hybridization
-
Labelling technique that uses the hybridization of labelled cDNA to locate specific nucleic acid sequences in tissue sections.
- Enzyme-linked immunosorbent assay
-
(ELISA). Plate-based antibody detection assay for biomolecules in which enzyme–antibody conjugates attach to specific antigens anchored to a surface and subsequent incubation in a substrate reveals the presence of antigens.
- Mass cytometry
-
(CyTOF). Variant of flow cytometry using metal ion-labelled antibodies and readout using time-of-flight mass spectrometry.
- Combinatorial indexing
-
Single-cell RNA sequencing method using transposase nuclei barcoding, fluorescence-activated nuclei sorting and PCR to index subpopulations of cells from tissues or organs.
Rights and permissions
About this article
Cite this article
Paik, D.T., Cho, S., Tian, L. et al. Single-cell RNA sequencing in cardiovascular development, disease and medicine. Nat Rev Cardiol 17, 457–473 (2020). https://doi.org/10.1038/s41569-020-0359-y
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41569-020-0359-y
This article is cited by
-
Intestinal cell diversity and treatment responses in a parasitic nematode at single cell resolution
BMC Genomics (2024)
-
A Multimodal Omics Framework to Empower Target Discovery for Cardiovascular Regeneration
Cardiovascular Drugs and Therapy (2024)
-
DCATS: differential composition analysis for flexible single-cell experimental designs
Genome Biology (2023)
-
Current and future perspectives of single-cell multi-omics technologies in cardiovascular research
Nature Cardiovascular Research (2023)
-
Pathway centric analysis for single-cell RNA-seq and spatial transcriptomics data with GSDensity
Nature Communications (2023)