Abstract
Advances in our understanding of how the gut microbiota contributes to human health and diseases have expanded our insight into how microbial composition and function affect the human host. Heart failure is associated with splanchnic circulation congestion, leading to bowel wall oedema and impaired intestinal barrier function. This situation is thought to heighten the overall inflammatory state via increased bacterial translocation and the presence of bacterial products in the systemic blood circulation. Several metabolites produced by gut microorganisms from dietary metabolism have been linked to pathologies such as atherosclerosis, hypertension, heart failure, chronic kidney disease, obesity, and type 2 diabetes mellitus. These findings suggest that the gut microbiome functions like an endocrine organ by generating bioactive metabolites that can directly or indirectly affect host physiology. In this Review, we discuss several newly discovered gut microbial metabolic pathways, including the production of trimethylamine and trimethylamine N-oxide, short-chain fatty acids, and secondary bile acids, that seem to participate in the development and progression of cardiovascular diseases, including heart failure. We also discuss the gut microbiome as a novel therapeutic target for the treatment of cardiovascular disease, and potential strategies for targeting intestinal microbial processes.
Key points
-
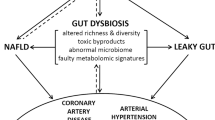
The gut hypothesis of heart failure (HF) postulates that bowel wall oedema and impaired intestinal barrier function in HF can promote bacterial translocation and inflammation.
-
Changes in the composition and diversity of the gut microbiota have been observed in patients with HF.
-
Microbial metabolites, especially those derived from dietary nutrients, can generate paracrine and endocrine effects that can lead to an increased susceptibility to HF.
-
Novel therapeutic strategies targeting gut microbial metabolic pathways and/or metabolites, as well as altering gut microbial composition, have the potential to modulate cardiovascular disease susceptibility and prevent progression to HF.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Yancy, C. W. et al. 2013 ACCF/AHA guideline for the management of heart failure: executive summary: a report of the American College of Cardiology Foundation/American Heart Association Task Force on practice guidelines. Circulation 128, 1810–1852 (2013).
Yancy, C. W. et al. 2017 ACC/AHA/HFSA focused update of the 2013 ACCF/AHA guideline for the management of heart failure: a report of the American College of Cardiology/American Heart Association Task Force on clinical practice guidelines and the Heart Failure Society of America. J. Am. Coll. Cardiol. 70, 776–803 (2017).
Anker, S. D. et al. Elevated soluble CD14 receptors and altered cytokines in chronic heart failure. Am. J. Cardiol. 79, 1426–1430 (1997).
Krack, A., Sharma, R., Figulla, H. R. & Anker, S. D. The importance of the gastrointestinal system in the pathogenesis of heart failure. Eur. Heart J. 26, 2368–2374 (2005).
Sender, R., Fuchs, S. & Milo, R. Revised estimates for the number of human and bacteria cells in the body. PLOS Biol. 14, e1002533 (2016).
Turnbaugh, P. J. et al. The human microbiome project. Nature 449, 804–810 (2007).
Dethlefsen, L., Eckburg, P. B., Bik, E. M. & Relman, D. A. Assembly of the human intestinal microbiota. Trends Ecol. Evol. 21, 517–523 (2006).
Human Microbiome Project Consortium. Structure, function and diversity of the healthy human microbiome. Nature 486, 207–214 (2012).
Booijink, C. C., Zoetendal, E. G., Kleerebezem, M. & de Vos, W. M. Microbial communities in the human small intestine: coupling diversity to metagenomics. Future Microbiol. 2, 285–295 (2007).
Gorbach, S. L. in Medical Microbiology 4th edn (ed. Baron, S.) (Univ. of Texas Medical Branch, 1996).
Hooper, L. V. et al. Molecular analysis of commensal host-microbial relationships in the intestine. Science 291, 881–884 (2001).
Marcobal, A. et al. Consumption of human milk oligosaccharides by gut-related microbes. J. Agr. Food Chem. 58, 5334–5340 (2010).
Belkaid, Y. & Hand, T. W. Role of the microbiota in immunity and inflammation. Cell 157, 121–141 (2014).
Fujiya, M. et al. The Bacillus subtilis quorum-sensing molecule CSF contributes to intestinal homeostasis via OCTN2, a host cell membrane transporter. Cell Host Microbe 1, 299–308 (2007).
Buffie, C. G. & Pamer, E. G. Microbiota-mediated colonization resistance against intestinal pathogens. Nat. Rev. Immunol. 13, 790–801 (2013).
Corr, S. C. et al. Bacteriocin production as a mechanism for the antiinfective activity of Lactobacillus salivarius UCC118. Proc. Natl Acad. Sci. USA 104, 7617–7621 (2007).
Rajilic-Stojanovic, M., Smidt, H. & de Vos, W. M. Diversity of the human gastrointestinal tract microbiota revisited. Environ. Microbiol. 9, 2125–2136 (2007).
Yang, X., Xie, L., Li, Y. & Wei, C. More than 9,000,000 unique genes in human gut bacterial community: estimating gene numbers inside a human body. PLOS ONE 4, e6074 (2009).
Palmer, C., Bik, E. M., DiGiulio, D. B., Relman, D. A. & Brown, P. O. Development of the human infant intestinal microbiota. PLOS Biol. 5, e177 (2007).
Dominguez-Bello, M. G. et al. Delivery mode shapes the acquisition and structure of the initial microbiota across multiple body habitats in newborns. Proc. Natl Acad. Sci. USA 107, 11971–11975 (2010).
Chu, D. M. et al. Maturation of the infant microbiome community structure and function across multiple body sites and in relation to mode of delivery. Nat. Med. 23, 314–326 (2017).
Hall, A. B., Tolonen, A. C. & Xavier, R. J. Human genetic variation and the gut microbiome in disease. Nat. Rev. Genet. 18, 690–699 (2017).
Koppel, N., Maini Rekdal, V. & Balskus, E. P. Chemical transformation of xenobiotics by the human gut microbiota. Science 356, eaag2770 (2017).
Sonnenburg, J. L. & Backhed, F. Diet-microbiota interactions as moderators of human metabolism. Nature 535, 56–64 (2016).
Brown, J. M. & Hazen, S. L. Targeting of microbe-derived metabolites to improve human health: the next frontier for drug discovery. J. Biol. Chem. 292, 8560–8568 (2017).
Saitta, K. S. et al. Bacterial beta-glucuronidase inhibition protects mice against enteropathy induced by indomethacin, ketoprofen or diclofenac: mode of action and pharmacokinetics. Xenobiotica 44, 28–35 (2014).
Wallace, B. D. et al. Structure and inhibition of microbiome β-glucuronidases essential to the alleviation of cancer drug toxicity. Chem. Biol. 22, 1238–1249 (2015).
Hermiston, M. L. & Gordon, J. I. Inflammatory bowel disease and adenomas in mice expressing a dominant negative N-cadherin. Science 270, 1203–1207 (1995).
Sandek, A. et al. Altered intestinal function in patients with chronic heart failure. J. Am. Coll. Cardiol. 50, 1561–1569 (2007).
Fink, M. P., Kaups, K. L., Wang, H. L. & Rothschild, H. R. Maintenance of superior mesenteric arterial perfusion prevents increased intestinal mucosal permeability in endotoxic pigs. Surgery 110, 154–160 (1991).
Triposkiadis, F. et al. The sympathetic nervous system in heart failure physiology, pathophysiology, and clinical implications. J. Am. Coll. Cardiol. 54, 1747–1762 (2009).
Jodal, M. & Lundgren, O. Countercurrent mechanisms in the mammalian gastrointestinal tract. Gastroenterology 91, 225–241 (1986).
Maynard, N. et al. Assessment of splanchnic oxygenation by gastric tonometry in patients with acute circulatory failure. JAMA 270, 1203–1210 (1993).
Krack, A. et al. Studies on intragastric PCO2 at rest and during exercise as a marker of intestinal perfusion in patients with chronic heart failure. Eur. J. Heart Fail. 6, 403–407 (2004).
Arutyunov, G. P., Kostyukevich, O. I., Serov, R. A., Rylova, N. V. & Bylova, N. A. Collagen accumulation and dysfunctional mucosal barrier of the small intestine in patients with chronic heart failure. Int. J. Cardiol. 125, 240–245 (2008).
Lu, Y. C., Yeh, W. C. & Ohashi, P. S. LPS/TLR4 signal transduction pathway. Cytokine 42, 145–151 (2008).
Frangogiannis, N. G. The inflammatory response in myocardial injury, repair, and remodelling. Nat. Rev. Cardiol. 11, 255–265 (2014).
Niebauer, J. et al. Endotoxin and immune activation in chronic heart failure: a prospective cohort study. Lancet 353, 1838–1842 (1999).
Peschel, T. et al. Invasive assessment of bacterial endotoxin and inflammatory cytokines in patients with acute heart failure. Eur. J. Heart Fail. 5, 609–614 (2003).
Hietbrink, F. et al. Systemic inflammation increases intestinal permeability during experimental human endotoxemia. Shock 32, 374–378 (2009).
Munger, M. A., Johnson, B., Amber, I. J., Callahan, K. S. & Gilbert, E. M. Circulating concentrations of proinflammatory cytokines in mild or moderate heart failure secondary to ischemic or idiopathic dilated cardiomyopathy. Am. J. Cardiol. 77, 723–727 (1996).
Paulus, W. J. How are cytokines activated in heart failure? Eur. J. Heart Fail. 1, 309–312 (1999).
Deswal, A. et al. Cytokines and cytokine receptors in advanced heart failure: an analysis of the cytokine database from the Vesnarinone trial (VEST). Circulation 103, 2055–2059 (2001).
Rauchhaus, M. et al. Plasma cytokine parameters and mortality in patients with chronic heart failure. Circulation 102, 3060–3067 (2000).
Ferrari, R. et al. Tumor necrosis factor soluble receptors in patients with various degrees of congestive heart failure. Circulation 92, 1479–1486 (1995).
Conraads, V. M. et al. Selective intestinal decontamination in advanced chronic heart failure: a pilot trial. Eur. J. Heart Fail. 6, 483–491 (2004).
Mann, D. L. Innate immunity and the failing heart: the cytokine hypothesis revisited. Circ. Res. 116, 1254–1268 (2015).
Bozkurt, B. et al. Pathophysiologically relevant concentrations of tumor necrosis factor-alpha promote progressive left ventricular dysfunction and remodeling in rats. Circulation 97, 1382–1391 (1998).
Kubota, T. et al. Dilated cardiomyopathy in transgenic mice with cardiac-specific overexpression of tumor necrosis factor-alpha. Circ. Res. 81, 627–635 (1997).
Al-Sadi, R. et al. Interleukin-6 modulation of intestinal epithelial tight junction permeability is mediated by JNK pathway activation of claudin-2 gene. PLOS ONE 9, e85345 (2014).
Al-Sadi, R. M. & Ma, T. Y. IL-1beta causes an increase in intestinal epithelial tight junction permeability. J. Immunol. 178, 4641–4649 (2007).
Ma, T. Y., Boivin, M. A., Ye, D., Pedram, A. & Said, H. M. Mechanism of TNF-{alpha} modulation of Caco-2 intestinal epithelial tight junction barrier: role of myosin light-chain kinase protein expression. Am. J. Physiol. Gastrointest. Liver Physiol. 288, G422–G430 (2005).
Al-Sadi, R., Guo, S., Ye, D. & Ma, T. Y. TNF-alpha modulation of intestinal epithelial tight junction barrier is regulated by ERK1/2 activation of Elk-1. Am. J. Pathol. 183, 1871–1884 (2013).
Chung, E. S. et al. Randomized, double-blind, placebo-controlled, pilot trial of infliximab, a chimeric monoclonal antibody to tumor necrosis factor-alpha, in patients with moderate-to-severe heart failure: results of the anti-TNF Therapy Against Congestive Heart Failure (ATTACH) trial. Circulation 107, 3133–3140 (2003).
Mann, D. L. et al. Targeted anticytokine therapy in patients with chronic heart failure: results of the Randomized Etanercept Worldwide Evaluation (RENEWAL). Circulation 109, 1594–1602 (2004).
Torre-Amione, G. et al. Results of a non-specific immunomodulation therapy in chronic heart failure (ACCLAIM trial): a placebo-controlled randomised trial. Lancet 371, 228–236 (2008).
Ridker, P. M. et al. Antiinflammatory therapy with canakinumab for atherosclerotic disease. N. Engl. J. Med. 377, 1119–1131 (2017).
Abbate, A. et al. Interleukin-1beta modulation using a genetically engineered antibody prevents adverse cardiac remodelling following acute myocardial infarction in the mouse. Eur. J. Heart Fail. 12, 319–322 (2010).
Abbate, A. et al. Anakinra, a recombinant human interleukin-1 receptor antagonist, inhibits apoptosis in experimental acute myocardial infarction. Circulation 117, 2670–2683 (2008).
Van Tassell, B. W. et al. Interleukin-1 trap attenuates cardiac remodeling after experimental acute myocardial infarction in mice. J. Cardiovasc. Pharmacol. 55, 117–122 (2010).
Van Tassell, B. W. et al. Interleukin-1 blockade in recently decompensated systolic heart failure: results from REDHART (Recently Decompensated Heart Failure Anakinra Response Trial). Circ. Heart Fail. 10, e004373 (2017).
Van Tassell, B. W. et al. Interleukin-1 blockade in heart failure with preserved ejection fraction: rationale and design of the Diastolic Heart Failure Anakinra Response Trial 2 (D-HART2). Clin. Cardiol. 40, 626–632 (2017).
US National Library of Medicine. ClinicalTrials.gov https://clinicaltrials.gov/ct2/show/NCT01900600 (2017).
Gullestad, L. et al. Inflammatory cytokines in heart failure: mediators and markers. Cardiology 122, 23–35 (2012).
Dick, S. A. & Epelman, S. Chronic heart failure and inflammation: what do we really know? Circ. Res. 119, 159–176 (2016).
Jakobsson, H. E. et al. The composition of the gut microbiota shapes the colon mucus barrier. EMBO Rep. 16, 164–177 (2015).
Tlaskalova-Hogenova, H. et al. The role of gut microbiota (commensal bacteria) and the mucosal barrier in the pathogenesis of inflammatory and autoimmune diseases and cancer: contribution of germ-free and gnotobiotic animal models of human diseases. Cell. Mol. Immunol. 8, 110–120 (2011).
Ley, R. E., Turnbaugh, P. J., Klein, S. & Gordon, J. I. Microbial ecology: human gut microbes associated with obesity. Nature 444, 1022–1023 (2006).
Turnbaugh, P. J. et al. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature 444, 1027–1031 (2006).
Morgan, X. C. et al. Dysfunction of the intestinal microbiome in inflammatory bowel disease and treatment. Genome Biol. 13, R79 (2012).
Wen, L. et al. Innate immunity and intestinal microbiota in the development of type 1 diabetes. Nature 455, 1109–1113 (2008).
Qin, J. et al. A metagenome-wide association study of gut microbiota in type 2 diabetes. Nature 490, 55–60 (2012).
Kummen, M. et al. The gut microbial profile in patients with primary sclerosing cholangitis is distinct from patients with ulcerative colitis without biliary disease and healthy controls. Gut 66, 611–619 (2017).
Sears, C. L. & Garrett, W. S. Microbes, microbiota, and colon cancer. Cell Host Microbe 15, 317–328 (2014).
Arthur, J. C. et al. Intestinal inflammation targets cancer-inducing activity of the microbiota. Science 338, 120–123 (2012).
Le Roy, T. et al. Intestinal microbiota determines development of non-alcoholic fatty liver disease in mice. Gut 62, 1787–1794 (2013).
Dumas, M. E. et al. Metabolic profiling reveals a contribution of gut microbiota to fatty liver phenotype in insulin-resistant mice. Proc. Natl Acad. Sci. USA 103, 12511–12516 (2006).
Hsiao, E. Y. et al. Microbiota modulate behavioral and physiological abnormalities associated with neurodevelopmental disorders. Cell 155, 1451–1463 (2013).
Foster, J. A. & McVey Neufeld, K. A. Gut-brain axis: how the microbiome influences anxiety and depression. Trends Neurosci. 36, 305–312 (2013).
Naseribafrouei, A. et al. Correlation between the human fecal microbiota and depression. Neurogastroenterol. Motil. 26, 1155–1162 (2014).
Gregory, J. C. et al. Transmission of atherosclerosis susceptibility with gut microbial transplantation. J. Biol. Chem. 290, 5647–5660 (2015).
Zhu, W. et al. Gut microbial metabolite TMAO enhances platelet hyperreactivity and thrombosis risk. Cell 165, 111–124 (2016).
Pluznick, J. L. et al. Olfactory receptor responding to gut microbiota-derived signals plays a role in renin secretion and blood pressure regulation. Proc. Natl Acad. Sci. USA 110, 4410–4415 (2013).
Mell, B. et al. Evidence for a link between gut microbiota and hypertension in the Dahl rat. Physiol. Genom. 47, 187–197 (2015).
Durgan, D. J. et al. Role of the gut microbiome in obstructive sleep apnea-induced hypertension. Hypertension 67, 469–474 (2016).
Lam, V. et al. Intestinal microbiota determine severity of myocardial infarction in rats. FASEB J. 26, 1727–1735 (2012).
Sandek, A. et al. Intestinal blood flow in patients with chronic heart failure: a link with bacterial growth, gastrointestinal symptoms, and cachexia. J. Am. Coll. Cardiol. 64, 1092–1102 (2014).
Pasini, E. et al. Pathogenic gut flora in patients with chronic heart failure. JACC Heart Fail. 4, 220–227 (2016).
Mamic, P., Heidenreich, P. A., Hedlin, H., Tennakoon, L. & Staudenmayer, K. L. Hospitalized patients with heart failure and common bacterial infections: a nationwide analysis of concomitant Clostridium difficile infection rates and in-hospital mortality. J. Card. Fail. 22, 891–900 (2016).
Luedde, M. et al. Heart failure is associated with depletion of core intestinal microbiota. ESC Heart Fail. 4, 282–290 (2017).
Cui, X. et al. Metagenomic and metabolomic analyses unveil dysbiosis of gut microbiota in chronic heart failure patients. Sci. Rep. 8, 635 (2018).
Kamo, T. et al. Dysbiosis and compositional alterations with aging in the gut microbiota of patients with heart failure. PLOS ONE 12, e0174099 (2017).
Kummen, M. et al. Gut microbiota signature in heart failure defined from profiling of 2 independent cohorts. J. Am. Coll. Cardiol. 71, 1184–1186 (2018).
Engels, C., Ruscheweyh, H. J., Beerenwinkel, N., Lacroix, C. & Schwab, C. The common gut microbe Eubacterium hallii also contributes to intestinal propionate formation. Front. Microbiol. 7, 713 (2016).
Karlsson, F. H. et al. Symptomatic atherosclerosis is associated with an altered gut metagenome. Nat. Commun. 3, 1245 (2012).
Wang, Z. et al. Non-lethal inhibition of gut microbial trimethylamine production for the treatment of atherosclerosis. Cell 163, 1585–1595 (2015).
Rothschild, D. et al. Environment dominates over host genetics in shaping human gut microbiota. Nature 555, 210–215 (2018).
Maier, L. et al. Extensive impact of non-antibiotic drugs on human gut bacteria. Nature 555, 623–628 (2018).
Caparros-Martin, J. A. et al. Statin therapy causes gut dysbiosis in mice through a PXR-dependent mechanism. Microbiome 5, 95 (2017).
Wu, H. et al. Metformin alters the gut microbiome of individuals with treatment-naive type 2 diabetes, contributing to the therapeutic effects of the drug. Nat. Med. 23, 850–858 (2017).
Haiser, H. J. et al. Predicting and manipulating cardiac drug inactivation by the human gut bacterium Eggerthella lenta. Science 341, 295–298 (2013).
Zhernakova, A. et al. Population-based metagenomics analysis reveals markers for gut microbiome composition and diversity. Science 352, 565–569 (2016).
Selwyn, F. P., Cui, J. Y. & Klaassen, C. D. RNA-seq quantification of hepatic drug processing genes in germ-free mice. Drug Metab. Dispos. 43, 1572–1580 (2015).
Bjorkholm, B. et al. Intestinal microbiota regulate xenobiotic metabolism in the liver. PLOS ONE 4, e6958 (2009).
Claus, S. P. et al. Colonization-induced host-gut microbial metabolic interaction. MBio 2, e00271 (2011).
Spanogiannopoulos, P., Bess, E. N., Carmody, R. N. & Turnbaugh, P. J. The microbial pharmacists within us: a metagenomic view of xenobiotic metabolism. Nat. Rev. Microbiol. 14, 273–287 (2016).
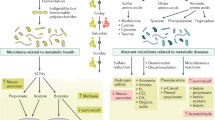
Topping, D. L. & Clifton, P. M. Short-chain fatty acids and human colonic function: roles of resistant starch and nonstarch polysaccharides. Physiol. Rev. 81, 1031–1064 (2001).
Hadjiagapiou, C., Schmidt, L., Dudeja, P. K., Layden, T. J. & Ramaswamy, K. Mechanism(s) of butyrate transport in Caco-2 cells: role of monocarboxylate transporter 1. Am. J. Physiol. Gastrointest. Liver Physiol. 279, G775–G780 (2000).
Mascolo, N., Rajendran, V. M. & Binder, H. J. Mechanism of short-chain fatty acid uptake by apical membrane vesicles of rat distal colon. Gastroenterology 101, 331–338 (1991).
Vidyasagar, S., Barmeyer, C., Geibel, J., Binder, H. J. & Rajendran, V. M. Role of short-chain fatty acids in colonic HCO(3) secretion. Am. J. Physiol. Gastrointest. Liver Physiol. 288, G1217–G1226 (2005).
den Besten, G. et al. The role of short-chain fatty acids in the interplay between diet, gut microbiota, and host energy metabolism. J. Lipid Res. 54, 2325–2340 (2013).
Sellin, J. H. SCFAs: the enigma of weak electrolyte transport in the colon. News Physiol. Sci. 14, 58–64 (1999).
Duncan, S. H., Louis, P., Thomson, J. M. & Flint, H. J. The role of pH in determining the species composition of the human colonic microbiota. Environ. Microbiol. 11, 2112–2122 (2009).
Cherrington, C. A., Hinton, M., Pearson, G. R. & Chopra, I. Short-chain organic acids at ph 5.0 kill Escherichia coli and Salmonella spp. without causing membrane perturbation. J. Appl. Bacteriol. 70, 161–165 (1991).
Pouteau, E., Meirim, I., Metairon, S. & Fay, L. B. Acetate, propionate and butyrate in plasma: determination of the concentration and isotopic enrichment by gas chromatography/mass spectrometry with positive chemical ionization. J. Mass Spectrom. 36, 798–805 (2001).
Yang, T. et al. Gut dysbiosis is linked to hypertension. Hypertension 65, 1331–1340 (2015).
Pluznick, J. A novel SCFA receptor, the microbiota, and blood pressure regulation. Gut Microbes 5, 202–207 (2014).
Marques, F. Z. et al. High-fiber diet and acetate supplementation change the gut microbiota and prevent the development of hypertension and heart failure in hypertensive mice. Circulation 135, 964–977 (2017).
Pouteau, E., Nguyen, P., Ballevre, O. & Krempf, M. Production rates and metabolism of short-chain fatty acids in the colon and whole body using stable isotopes. Proc. Nutr. Soc. 62, 87–93 (2003).
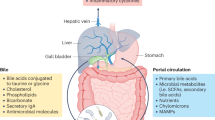
Lefebvre, P., Cariou, B., Lien, F., Kuipers, F. & Staels, B. Role of bile acids and bile acid receptors in metabolic regulation. Physiol. Rev. 89, 147–191 (2009).
Dawson, P. A., Lan, T. & Rao, A. Bile acid transporters. J. Lipid Res. 50, 2340–2357 (2009).
Devlin, A. S. & Fischbach, M. A. A biosynthetic pathway for a prominent class of microbiota-derived bile acids. Nat. Chem. Biol. 11, 685–690 (2015).
Ridlon, J. M., Kang, D. J. & Hylemon, P. B. Bile salt biotransformations by human intestinal bacteria. J. Lipid Res. 47, 241–259 (2006).
Hamilton, J. P. et al. Human cecal bile acids: concentration and spectrum. Am. J. Physiol. Gastrointest. Liver Physiol. 293, G256–G263 (2007).
Mayerhofer, C. C. K. et al. Increased secondary/primary bile acid ratio in chronic heart failure. J. Card. Fail. 23, 666–671 (2017).
Binah, O., Rubinstein, I., Bomzon, A. & Better, O. S. Effects of bile acids on ventricular muscle contraction and electrophysiological properties: studies in rat papillary muscle and isolated ventricular myocytes. Naunyn Schmiedebergs Arch. Pharmacol. 335, 160–165 (1987).
Joubert, P. An in vivo investigation of the negative chronotropic effect of cholic acid in the rat. Clin. Exp. Pharmacol. Physiol. 5, 1–8 (1978).
Gazawi, H. et al. The effects of bile acids on beta-adrenoceptors, fluidity, and the extent of lipid peroxidation in rat cardiac membranes. Biochem. Pharmacol. 59, 1623–1628 (2000).
Wang, H., Chen, J., Hollister, K., Sowers, L. C. & Forman, B. M. Endogenous bile acids are ligands for the nuclear receptor FXR/BAR. Mol. Cell 3, 543–553 (1999).
Forman, B. M. et al. Identification of a nuclear receptor that is activated by farnesol metabolites. Cell 81, 687–693 (1995).
Kawamata, Y. et al. A G protein-coupled receptor responsive to bile acids. J. Biol. Chem. 278, 9435–9440 (2003).
Rosen, H., Gonzalez-Cabrera, P. J., Sanna, M. G. & Brown, S. Sphingosine 1-phosphate receptor signaling. Annu. Rev. Biochem. 78, 743–768 (2009).
Raufman, J. P., Chen, Y., Zimniak, P. & Cheng, K. Deoxycholic acid conjugates are muscarinic cholinergic receptor antagonists. Pharmacology 65, 215–221 (2002).
Calkin, A. C. & Tontonoz, P. Transcriptional integration of metabolism by the nuclear sterol-activated receptors LXR and FXR. Nat. Rev. Mol. Cell Biol. 13, 213–224 (2012).
Sayin, S. I. et al. Gut microbiota regulates bile acid metabolism by reducing the levels of tauro-beta-muricholic acid, a naturally occurring FXR antagonist. Cell Metab. 17, 225–235 (2013).
Wang, Y. D. et al. Farnesoid X receptor antagonizes nuclear factor kappaB in hepatic inflammatory response. Hepatology 48, 1632–1643 (2008).
Li, Y. T., Swales, K. E., Thomas, G. J., Warner, T. D. & Bishop-Bailey, D. Farnesoid x receptor ligands inhibit vascular smooth muscle cell inflammation and migration. Arterioscler. Thromb. Vasc. Biol. 27, 2606–2611 (2007).
Purcell, N. H. et al. Activation of NF-kappa B is required for hypertrophic growth of primary rat neonatal ventricular cardiomyocytes. Proc. Natl Acad. Sci. USA 98, 6668–6673 (2001).
Gordon, J. W., Shaw, J. A. & Kirshenbaum, L. A. Multiple facets of NF-kappaB in the heart: to be or not to NF-kappaB. Circ. Res. 108, 1122–1132 (2011).
Pu, J. et al. Cardiomyocyte-expressed farnesoid-X-receptor is a novel apoptosis mediator and contributes to myocardial ischaemia/reperfusion injury. Eur. Heart J. 34, 1834–1845 (2013).
Ali, A. H., Carey, E. J. & Lindor, K. D. Recent advances in the development of farnesoid X receptor agonists. Ann. Transl Med. 3, 5 (2015).
Pellicciari, R. et al. 6α-ethyl-chenodeoxycholic acid (6-ECDCA), a potent and selective FXR agonist endowed with anticholestatic activity. J. Med. Chem. 45, 3569–3572 (2002).
Nevens, F. et al. A placebo-controlled trial of obeticholic acid in primary biliary cholangitis. N. Engl. J. Med. 375, 631–643 (2016).
Neuschwander-Tetri, B. A. et al. Farnesoid X nuclear receptor ligand obeticholic acid for non-cirrhotic, non-alcoholic steatohepatitis (FLINT): a multicentre, randomised, placebo-controlled trial. Lancet 385, 956–965 (2015).
Mudaliar, S. et al. Efficacy and safety of the farnesoid X receptor agonist obeticholic acid in patients with type 2 diabetes and nonalcoholic fatty liver disease. Gastroenterology 145, 574–582 (2013).
Xu, Y. et al. Farnesoid X receptor activation increases reverse cholesterol transport by modulating bile acid composition and cholesterol absorption in mice. Hepatology 64, 1072–1085 (2016).
Cipriani, S., Mencarelli, A., Palladino, G. & Fiorucci, S. FXR activation reverses insulin resistance and lipid abnormalities and protects against liver steatosis in Zucker (fa/fa) obese rats. J. Lipid Res. 51, 771–784 (2010).
von Haehling, S. et al. Ursodeoxycholic acid in patients with chronic heart failure: a double-blind, randomized, placebo-controlled, crossover trial. J. Am. Coll. Cardiol. 59, 585–592 (2012).
Aouad, K. et al. Immunosuppressive effects of endotoxins and bile acids in vivo in the rat. Eur. J. Clin. Invest. 26, 45–48 (1996).
Koeth, R. A. et al. Intestinal microbiota metabolism of L-carnitine, a nutrient in red meat, promotes atherosclerosis. Nat. Med. 19, 576–585 (2013).
Tang, W. H. et al. Intestinal microbial metabolism of phosphatidylcholine and cardiovascular risk. N. Engl. J. Med. 368, 1575–1584 (2013).
Wang, Z. et al. Gut flora metabolism of phosphatidylcholine promotes cardiovascular disease. Nature 472, 57–63 (2011).
Zhao, Y. et al. The effect of different L-carnitine administration routes on the development of atherosclerosis in ApoE knockout mice. Mol. Nutr. Food Res. 62, 1700299 (2018).
Organ, C. L. et al. Choline diet and its gut microbe-derived metabolite, trimethylamine N-oxide, exacerbate pressure overload-induced heart failure. Circ. Heart Fail. 9, e002314 (2016).
Seibel, B. A. & Walsh, P. J. Trimethylamine oxide accumulation in marine animals: relationship to acylglycerol storage. J. Exp. Biol. 205, 297–306 (2002).
Parab, S. N. & Rao, S. B. Distribution of trimethylamine oxie in some marine and freshwater fish. Curr. Sci. 53, 307–309 (1984).
Cladis, D. P., Kleiner, A. C., Freiser, H. H. & Santerre, C. R. Fatty acid profiles of commercially available finfish fillets in the United States. Lipids 49, 1005–1018 (2014).
Weaver, K. L. et al. The content of favorable and unfavorable polyunsaturated fatty acids found in commonly eaten fish. J. Am. Diet. Assoc. 108, 1178–1185 (2008).
Svensson, B. G., Akesson, B., Nilsson, A. & Paulsson, K. Urinary excretion of methylamines in men with varying intake of fish from the Baltic Sea. J. Toxicol. Environ. Health 41, 411–420 (1994).
Yazdekhasti, N. et al. Fish protein increases circulating levels of trimethylamine-N-oxide and accelerates aortic lesion formation in apoE null mice. Mol. Nutr. Food Res. 60, 358–368 (2016).
Gao, X. et al. Fish oil ameliorates trimethylamine N-oxide-exacerbated glucose intolerance in high-fat diet-fed mice. Food Funct. 6, 1117–1125 (2015).
Kasiske, B. L., Lakatua, J. D., Ma, J. Z. & Louis, T. A. A meta-analysis of the effects of dietary protein restriction on the rate of decline in renal function. Am. J. Kidney Dis. 31, 954–961 (1998).
Pedrini, M. T., Levey, A. S., Lau, J., Chalmers, T. C. & Wang, P. H. The effect of dietary protein restriction on the progression of diabetic and nondiabetic renal diseases: a meta-analysis. Ann. Intern. Med. 124, 627–632 (1996).
Collins, H. L. et al. L-carnitine intake and high trimethylamine N-oxide plasma levels correlate with low aortic lesions in ApoE −/− transgenic mice expressing CETP. Atherosclerosis 244, 29–37 (2016).
Bennett, B. J. et al. Trimethylamine-N-oxide, a metabolite associated with atherosclerosis, exhibits complex genetic and dietary regulation. Cell Metab. 17, 49–60 (2013).
Zhu, W., Wang, Z., Tang, W. H. W. & Hazen, S. L. Gut microbe-generated trimethylamine N-oxide from dietary choline is prothrombotic in subjects. Circulation 135, 1671–1673 (2017).
Seldin, M. M. et al. Trimethylamine N-oxide promotes vascular inflammation through signaling of mitogen-activated protein kinase and nuclear factor-kappaB. J. Am. Heart Assoc. 5, e002767 (2016).
Sun, X. et al. Trimethylamine N-oxide induces inflammation and endothelial dysfunction in human umbilical vein endothelial cells via activating ROS-TXNIP-NLRP3 inflammasome. Biochem. Biophys. Res. Commun. 481, 63–70 (2016).
Ma, G. et al. Trimethylamine N-oxide in atherogenesis: impairing endothelial self-repair capacity and enhancing monocyte adhesion. Biosci. Rep. 37, BSR20160244 (2017).
Bennion, B. J. & Daggett, V. Counteraction of urea-induced protein denaturation by trimethylamine N-oxide: a chemical chaperone at atomic resolution. Proc. Natl Acad. Sci. USA 101, 6433–6438 (2004).
Liberles, S. D. & Buck, L. B. A second class of chemosensory receptors in the olfactory epithelium. Nature 442, 645–650 (2006).
Ufnal, M. et al. Trimethylamine-N-oxide: a carnitine-derived metabolite that prolongs the hypertensive effect of angiotensin II in rats. Can. J. Cardiol. 30, 1700–1705 (2014).
Organ, C. L. et al. Removal of dietary trimethylamine N-oxide (TMAO) attenuates cardiac dysfunction in pressure overload induced heart failure [abstract]. Circulation 134 (Suppl. 1), 19293 (2016).
Chen, K., Zheng, X., Feng, M., Li, D. & Zhang, H. Gut microbiota-dependent metabolite trimethylamine N-oxide contributes to cardiac dysfunction in western diet-induced obese mice. Front. Physiol. 8, 139 (2017).
Tang, W. H. et al. Prognostic value of elevated levels of intestinal microbe-generated metabolite trimethylamine-N-oxide in patients with heart failure: refining the gut hypothesis. J. Am. Coll. Cardiol. 64, 1908–1914 (2014).
Tang, W. H. et al. Intestinal microbiota-dependent phosphatidylcholine metabolites, diastolic dysfunction, and adverse clinical outcomes in chronic systolic heart failure. J. Card. Fail. 21, 91–96 (2015).
Troseid, M. et al. Microbiota-dependent metabolite trimethylamine-N-oxide is associated with disease severity and survival of patients with chronic heart failure. J. Intern. Med. 277, 717–726 (2015).
Schuett, K. et al. Trimethylamine-N-oxide and heart failure with reduced versus preserved ejection fraction. J. Am. Coll. Cardiol. 70, 3202–3204 (2017).
Suzuki, T., Heaney, L. M., Bhandari, S. S., Jones, D. J. & Ng, L. L. Trimethylamine N-oxide and prognosis in acute heart failure. Heart 102, 841–848 (2016).
Savi, M. et al. Trimethylamine-N-oxide (TMAO)-induced impairment of cardiomyocyte function and the protective role of urolithin B-glucuronide. Molecules 23, E549 (2018).
Oakley, C. et al. The uremic metabolite, trimethylamine-N-oxide (TMAO), directly alters cardiac contractile function [abstract]. FASEB J. 31 (Suppl. 1), 688.3 (2017).
Schefold, J. C., Filippatos, G., Hasenfuss, G., Anker, S. D. & von Haehling, S. Heart failure and kidney dysfunction: epidemiology, mechanisms and management. Nat. Rev. Nephrol. 12, 610–623 (2016).
Hillege, H. L. et al. Renal function as a predictor of outcome in a broad spectrum of patients with heart failure. Circulation 113, 671–678 (2006).
van Deursen, V. M. et al. Co-morbidities in patients with heart failure: an analysis of the European Heart Failure Pilot Survey. Eur. J. Heart Fail. 16, 103–111 (2014).
Cho, C. E. et al. Trimethylamine-N-oxide (TMAO) response to animal source foods varies among healthy young men and is influenced by their gut microbiota composition: a randomized controlled trial. Mol. Nutr. Food Res. 61, 1600324 (2017).
Smith, J. L., Wishnok, J. S. & Deen, W. M. Metabolism and excretion of methylamines in rats. Toxicol. Appl. Pharmacol. 125, 296–308 (1994).
Forster, R. P., Berglund, F. & Rennick, B. R. Tubular secretion of creatine, trimethylamine oxide, and other organic bases by the aglomerular kidney of Lophius americanus. J. Gen. Physiol. 42, 319–327 (1958).
Cohen, J. J., Krupp, M. A., Chidsey, C. A. 3rd & Biltz, C. I. Effect of trimethylamine and its homologues on renal conservation of trimethylamine oxide by the spiny dogfish, Squalus acanthias. Am. J. Physiol. 196, 93–99 (1959).
Acara, M., Camiolo, S. & Rennick, B. Renal N-oxidation of trimethylamine in the chicken during tubular excretion. Drug Metab. Dispos. 5, 82–90 (1977).
Bell, J. D. et al. Nuclear magnetic resonance studies of blood plasma and urine from subjects with chronic renal failure: identification of trimethylamine-N-oxide. Biochim. Biophys. Acta 1096, 101–107 (1991).
Rhee, E. P. et al. A combined epidemiologic and metabolomic approach improves CKD prediction. J. Am. Soc. Nephrol. 24, 1330–1338 (2013).
Tang, W. H. et al. Gut microbiota-dependent trimethylamine N-oxide (TMAO) pathway contributes to both development of renal insufficiency and mortality risk in chronic kidney disease. Circ. Res. 116, 448–455 (2015).
Stubbs, J. R. et al. Serum trimethylamine-N-oxide is elevated in CKD and correlates with coronary atherosclerosis burden. J. Am. Soc. Nephrol. 27, 305–313 (2016).
Kim, R. B. et al. Advanced chronic kidney disease populations have elevated trimethylamine N-oxide levels associated with increased cardiovascular events. Kidney Int. 89, 1144–1152 (2016).
Shafi, T. et al. Trimethylamine N-oxide and cardiovascular events in hemodialysis patients. J. Am. Soc. Nephrol. 28, 321–331 (2017).
Missailidis, C. et al. Serum trimethylamine-N-oxide is strongly related to renal function and predicts outcome in chronic kidney disease. PLOS ONE 11, e0141738 (2016).
Gruppen, E. G. et al. TMAO is associated with mortality: impact of modestly impaired renal function. Sci. Rep. 7, 13781 (2017).
Hartiala, J. et al. Comparative genome-wide association studies in mice and humans for trimethylamine N-oxide, a proatherogenic metabolite of choline and L-carnitine. Arterioscler. Thromb. Vasc. Biol. 34, 1307–1313 (2014).
Robinson-Cohen, C. et al. Association of FMO3 variants and trimethylamine N-oxide concentration, disease progression, and mortality in CKD patients. PLOS ONE 11, e0161074 (2016).
Johnson, C., Prokopienko, A. J., West, R. E. 3rd, Nolin, T. D. & Stubbs, J. R. Decreased kidney function is associated with enhanced hepatic flavin monooxygenase activity and increased circulating trimethylamine N-oxide concentrations in mice. Drug Metab. Dispos. 46, 1304–1309 (2018).
Heianza, Y., Ma, W., Manson, J. E., Rexrode, K. M. & Qi, L. Gut microbiota metabolites and risk of major adverse cardiovascular disease events and death: a systematic review and meta-analysis of prospective studies. J. Am. Heart Assoc. 6, e004947 (2017).
Qi, J. et al. Circulating trimethylamine N-oxide and the risk of cardiovascular diseases: a systematic review and meta-analysis of 11 prospective cohort studies. J. Cell. Mol. Med. 22, 185–194 (2017).
Schiattarella, G. G. et al. Gut microbe-generated metabolite trimethylamine-N-oxide as cardiovascular risk biomarker: a systematic review and dose-response meta-analysis. Eur. Heart J. 38, 2948–2956 (2017).
Gan, X. T. et al. Probiotic administration attenuates myocardial hypertrophy and heart failure after myocardial infarction in the rat. Circ. Heart Fail. 7, 491–499 (2014).
Costanza, A. C., Moscavitch, S. D., Faria Neto, H. C. & Mesquita, E. T. Probiotic therapy with Saccharomyces boulardii for heart failure patients: a randomized, double-blind, placebo-controlled pilot trial. Int. J. Cardiol. 179, 348–350 (2015).
Kochan, P. et al. Lactobacillus rhamnosus administration causes sepsis in a cardiosurgical patient—is the time right to revise probiotic safety guidelines? Clin. Microbiol. Infect. 17, 1589–1592 (2011).
Liong, M. T. Safety of probiotics: translocation and infection. Nutr. Rev. 66, 192–202 (2008).
Salehi-Abargouei, A., Maghsoudi, Z., Shirani, F. & Azadbakht, L. Effects of dietary approaches to stop hypertension (DASH)-style diet on fatal or nonfatal cardiovascular diseases—incidence: a systematic review and meta-analysis on observational prospective studies. Nutrition 29, 611–618 (2013).
Levitan, E. B., Wolk, A. & Mittleman, M. A. Consistency with the DASH diet and incidence of heart failure. Arch. Intern. Med. 169, 851–857 (2009).
Levitan, E. B., Wolk, A. & Mittleman, M. A. Relation of consistency with the dietary approaches to stop hypertension diet and incidence of heart failure in men aged 45 to 79 years. Am. J. Cardiol. 104, 1416–1420 (2009).
Nguyen, H. T. et al. DASH eating pattern is associated with favorable left ventricular function in the multi-ethnic study of atherosclerosis. J. Am. Coll. Nutr. 31, 401–407 (2012).
Rifai, L., Pisano, C., Hayden, J., Sulo, S. & Silver, M. A. Impact of the DASH diet on endothelial function, exercise capacity, and quality of life in patients with heart failure. Proc. (Bayl Univ. Med. Cent.) 28, 151–156 (2015).
Mathew, A. V., Seymour, E. M., Byun, J., Pennathur, S. & Hummel, S. L. Altered metabolic profile with sodium-restricted dietary approaches to stop hypertension diet in hypertensive heart failure with preserved ejection fraction. J. Card. Fail. 21, 963–967 (2015).
Hummel, S. L. et al. Low-sodium DASH diet improves diastolic function and ventricular-arterial coupling in hypertensive heart failure with preserved ejection fraction. Circ. Heart Fail. 6, 1165–1171 (2013).
Hummel, S. L. et al. Low-sodium dietary approaches to stop hypertension diet reduces blood pressure, arterial stiffness, and oxidative stress in hypertensive heart failure with preserved ejection fraction. Hypertension 60, 1200–1206 (2012).
Estruch, R. et al. Primary prevention of cardiovascular disease with a mediterranean diet supplemented with extra-virgin olive oil or nuts. N. Engl. J. Med. 378, e34 (2018).
Papadaki, A. et al. Mediterranean diet and risk of heart failure: results from the PREDIMED randomized controlled trial. Eur. J. Heart Fail. 19, 1179–1185 (2017).
Miro, O. et al. Adherence to mediterranean diet and all-cause mortality after an episode of acute heart failure: results of the MEDIT-AHF study. JACC Heart Fail. 6, 52–62 (2018).
Falony, G., Vieira-Silva, S. & Raes, J. Microbiology meets big data: the case of gut microbiota-derived trimethylamine. Annu. Rev. Microbiol. 69, 305–321 (2015).
Craciun, S. & Balskus, E. P. Microbial conversion of choline to trimethylamine requires a glycyl radical enzyme. Proc. Natl Acad. Sci. USA 109, 21307–21312 (2012).
Koeth, R. A. et al. γ-Butyrobetaine is a proatherogenic intermediate in gut microbial metabolism of L-carnitine to TMAO. Cell Metab. 20, 799–812 (2014).
Meyer, M., Granderath, K. & Andreesen, J. R. Purification and characterization of protein PB of betaine reductase and its relationship to the corresponding proteins glycine reductase and sarcosine reductase from Eubacterium acidaminophilum. Eur. J. Biochem. 234, 184–191 (1995).
Wagner, M. et al. Substrate-specific selenoprotein B of glycine reductase from Eubacterium acidaminophilum. Biochemical and molecular analysis. Eur. J. Biochem. 260, 38–49 (1999).
Zhu, Y. et al. Carnitine metabolism to trimethylamine by an unusual Rieske-type oxygenase from human microbiota. Proc. Natl Acad. Sci. USA 111, 4268–4273 (2014).
Kuka, J. et al. Suppression of intestinal microbiota-dependent production of pro-atherogenic trimethylamine N-oxide by shifting L-carnitine microbial degradation. Life Sci. 117, 84–92 (2014).
Chen, M. L. et al. Resveratrol attenuates trimethylamine-N-oxide (TMAO)-induced atherosclerosis by regulating TMAO synthesis and bile acid metabolism via remodeling of the gut microbiota. MBio 7, e02210 (2016).
Johnson, E. L., Heaver, S. L., Walters, W. A. & Ley, R. E. Microbiome and metabolic disease: revisiting the bacterial phylum Bacteroidetes. J. Mol. Med. 95, 1–8 (2017).
Arumugam, M. et al. Enterotypes of the human gut microbiome. Nature 473, 174–180 (2011).
Jeffery, I. B., Claesson, M. J., O’Toole, P. W. & Shanahan, F. Categorization of the gut microbiota: enterotypes or gradients? Nat. Rev. Microbiol. 10, 591–592 (2012).
Knights, D. et al. Rethinking “enterotypes”. Cell Host Microbe 16, 433–437 (2014).
Li, J. et al. An integrated catalog of reference genes in the human gut microbiome. Nat. Biotechnol. 32, 834–841 (2014).
Martinez-del Campo, A. et al. Characterization and detection of a widely distributed gene cluster that predicts anaerobic choline utilization by human gut bacteria. MBio 6, e00042 (2015).
Hur, K. Y. & Lee, M. S. Gut microbiota and metabolic disorders. Diabetes Metab. J. 39, 198–203 (2015).
Acknowledgements
The authors’ research work is supported by grants from the National Institutes of Health (NIH) and the Office of Dietary Supplements, including R01HL103866 (S.L.H.), and P20HL113452, R01DK106000, and R01HL126827 (W.H.W.T. and S.L.H.).
Author information
Authors and Affiliations
Contributions
W.H.W.T. and D.Y.L. researched data for the article, and all authors discussed its content. W.H.W.T. and D.Y.L. wrote the manuscript, and W.H.W.T. and S.L.H. reviewed and edited it before submission.
Corresponding author
Ethics declarations
Competing interests
S.L.H. is named as a co-inventor on pending and issued patents held by the Cleveland Clinic, USA, relating to cardiovascular diagnostics and therapeutics; he is a paid consultant for Proctor & Gamble and has received research funds from Proctor & Gamble and Roche Diagnostics; and he is eligible to receive royalty payments for inventions or discoveries related to cardiovascular diagnostics or therapeutics from Cleveland Heart Lab/Quest Diagnostics or Proctor & Gamble. The other authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Related links
NIH Human Microbiome Project: https://hmpdacc.org/
Metagenomics of the Human Intestinal Tract (MetaHIT): http://www.metahit.eu/
Glossary
- Meta-organismal
-
Relating to a polygenomic organism consisting of a community of living beings of different species (such as microbiota and host) associated by interspecies symbiosis.
Rights and permissions
About this article
Cite this article
Tang, W.H.W., Li, D.Y. & Hazen, S.L. Dietary metabolism, the gut microbiome, and heart failure. Nat Rev Cardiol 16, 137–154 (2019). https://doi.org/10.1038/s41569-018-0108-7
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41569-018-0108-7
This article is cited by
-
Quinic acid regulated TMA/TMAO-related lipid metabolism and vascular endothelial function through gut microbiota to inhibit atherosclerotic
Journal of Translational Medicine (2024)
-
Microbes little helpers and suppliers for therapeutic asthma approaches
Respiratory Research (2024)
-
Gut Microbiome and Polycystic Ovary Syndrome: Interplay of Associated Microbial-Metabolite Pathways and Therapeutic Strategies
Reproductive Sciences (2024)
-
Kidney damage relates to agonal bacteremia: a single-center retrospective study
Clinical and Experimental Nephrology (2024)
-
Heart Failure: a Punch from the Gut
Current Heart Failure Reports (2024)