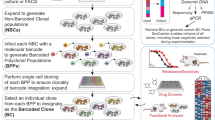
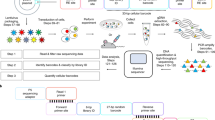
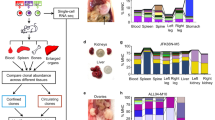
Abstract
Tumours are often composed of a multitude of malignant clones that are genomically unique, and only a few of them may have the ability to escape cancer therapy and grow as symptomatic lesions. As a result, tumours with a large degree of genomic diversity have a higher chance of leading to patient death. However, clonal fate can be driven by non-genomic features. In this context, new technologies are emerging not only to track the spatiotemporal fate of individual cells and their progeny but also to study their molecular features using various omics analysis. In particular, the recent development of cellular barcoding facilitates the labelling of tens to millions of cancer clones and enables the identification of the complex mechanisms associated with clonal fate in different microenvironments and in response to therapy. In this Review, we highlight the recent discoveries made using lentiviral-based cellular barcoding techniques, namely genetic and optical barcoding. We also emphasize the strengths and limitations of each of these technologies and discuss some of the key concepts that must be taken into consideration when one is designing barcoding experiments. Finally, we suggest new directions to further improve the use of these technologies in cancer research.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
References
Marusyk, A., Janiszewska, M. & Polyak, K. Intratumor heterogeneity: the Rosetta stone of therapy resistance. Cancer Cell 37, 471–484 (2020).
Shembrey, C., Huntington, N. D. & Hollande, F. Impact of tumor and immunological heterogeneity on the anti-cancer immune response. Cancers https://doi.org/10.3390/cancers11091217 (2019).
Tabassum, D. P. & Polyak, K. Tumorigenesis: it takes a village. Nat. Rev. Cancer 15, 473–483 (2015).
Curtis, C. et al. The genomic and transcriptomic architecture of 2,000 breast tumours reveals novel subgroups. Nature 486, 346–352 (2012).
Kim, C. et al. Chemoresistance evolution in triple-negative breast cancer delineated by single-cell sequencing. Cell 173, 879–893 e813 (2018).
Jia, Q., Chu, H., Jin, Z., Long, H. & Zhu, B. High-throughput single-cell sequencing in cancer research. Signal. Transduct. Target. Ther. 7, 145 (2022).
Nam, A. S., Chaligne, R. & Landau, D. A. Integrating genetic and non-genetic determinants of cancer evolution by single-cell multi-omics. Nat. Rev. Genet. 22, 3–18 (2021).
Merino, D. et al. Barcoding reveals complex clonal behavior in patient-derived xenografts of metastatic triple negative breast cancer. Nat. Commun. 10, 766 (2019).
Naik, S. H., Schumacher, T. N. & Perie, L. Cellular barcoding: a technical appraisal. Exp. Hematol. 42, 598–608 (2014).
Bystrykh, L. V. & Belderbos, M. E. Clonal analysis of cells with cellular barcoding: when numbers and sizes matter. Methods Mol. Biol. 1516, 57–89 (2016).
Kebschull, J. M. & Zador, A. M. Cellular barcoding: lineage tracing, screening and beyond. Nat. Methods 15, 871–879 (2018).
Lewis, S. M. et al. Spatial omics and multiplexed imaging to explore cancer biology. Nat. Methods https://doi.org/10.1038/s41592-021-01203-6 (2021).
Gui, P. & Bivona, T. G. Evolution of metastasis: new tools and insights. Trends Cancer https://doi.org/10.1016/j.trecan.2021.11.002 (2021).
Klein, C. A. Parallel progression of primary tumours and metastases. Nat. Rev. Cancer 9, 302–312 (2009).
Dagogo-Jack, I. & Shaw, A. T. Tumour heterogeneity and resistance to cancer therapies. Nat. Rev. Clin. Oncol. 15, 81–94 (2018).
Livet, J. et al. Transgenic strategies for combinatorial expression of fluorescent proteins in the nervous system. Nature 450, 56–62 (2007).
Snippert, H. J. et al. Intestinal crypt homeostasis results from neutral competition between symmetrically dividing Lgr5 stem cells. Cell 143, 134–144 (2010).
Dumas, L., Clavreul, S., Michon, F. & Loulier, K. Multicolor strategies for investigating clonal expansion and tissue plasticity. Cell Mol. Life Sci. 79, 141 (2022).
Baron, C. S. & van Oudenaarden, A. Unravelling cellular relationships during development and regeneration using genetic lineage tracing. Nat. Rev. Mol. Cell Biol. 20, 753–765 (2019).
McKenna, A. & Gagnon, J. A. Recording development with single cell dynamic lineage tracing. Development https://doi.org/10.1242/dev.169730 (2019).
Wagner, D. E. & Klein, A. M. Lineage tracing meets single-cell omics: opportunities and challenges. Nat. Rev. Genet. 21, 410–427 (2020).
Weber, K. et al. RGB marking facilitates multicolor clonal cell tracking. Nat. Med. 17, 504–509 (2011).
Coffey, S. E., Giedt, R. J. & Weissleder, R. Automated analysis of clonal cancer cells by intravital imaging. Intravital https://doi.org/10.4161/intv.26138 (2013).
Caiado, F. et al. Lineage tracing of acute myeloid leukemia reveals the impact of hypomethylating agents on chemoresistance selection. Nat. Commun. 10, 4986 (2019).
Berthelet, J. et al. The site of breast cancer metastases dictates their clonal composition and reversible transcriptomic profile. Sci. Adv. https://doi.org/10.1126/sciadv.abf4408 (2021). This study uses optical barcoding to map clonal fate and interactions of 31 breast cancer clones in vitro and in vivo, in multiple organs and in response to targeted therapy.
Nguyen, L. V. et al. DNA barcoding reveals diverse growth kinetics of human breast tumour subclones in serially passaged xenografts. Nat. Commun. 5, 5871 (2014). This study is one of the first to use genetic barcoding to investigate the frequency of tumour-initiating cells in retransplantation experiments using PDXs.
Naik, S. H. et al. Diverse and heritable lineage imprinting of early haematopoietic progenitors. Nature 496, 229–232 (2013).
Jordan, C. T. & Lemischka, I. R. Clonal and systemic analysis of long-term hematopoiesis in the mouse. Genes Dev. 4, 220–232 (1990).
Walsh, C. & Cepko, C. L. Widespread dispersion of neuronal clones across functional regions of the cerebral cortex. Science 255, 434–440 (1992).
Elder, A. et al. Abundant and equipotent founder cells establish and maintain acute lymphoblastic leukaemia. Leukemia 31, 2577–2586 (2017).
Nguyen, L. V. et al. Clonal analysis via barcoding reveals diverse growth and differentiation of transplanted mouse and human mammary stem cells. Cell Stem Cell 14, 253–263 (2014).
Seth, S. et al. Pre-existing functional heterogeneity of tumorigenic compartment as the origin of chemoresistance in pancreatic tumors. Cell Rep. 26, 1518–1532 e1519 (2019).
Fennell, K. A. et al. Non-genetic determinants of malignant clonal fitness at single-cell resolution. Nature https://doi.org/10.1038/s41586-021-04206-7 (2021). This article describes the generation of a high-diversity transcribed library, which is used to study, over time, the non-genetic processes influencing clonal fitness in leukaemia.
Oren, Y. et al. Cycling cancer persister cells arise from lineages with distinct programs. Nature https://doi.org/10.1038/s41586-021-03796-6 (2021). This study describes the development of the Watermelon library, which enables the characterization of drug-resistant clones at single-cell resolution.
Biddy, B. A. et al. Single-cell mapping of lineage and identity in direct reprogramming. Nature 564, 219–224 (2018).
Gutierrez, C. et al. Multifunctional barcoding with ClonMapper enables high-resolution study of clonal dynamics during tumor evolution and treatment. Nat. Cancer 2, 758–772 (2021). This study describes the use of a multifunctional library, ClonMapper, to combine lineage tracing, single-cell analysis and lineage recall to study the behaviour of leukaemia clones in response to chemotherapy.
Weinreb, C., Rodriguez-Fraticelli, A., Camargo, F. D. & Klein, A. M. Lineage tracing on transcriptional landscapes links state to fate during differentiation. Science https://doi.org/10.1126/science.aaw3381 (2020).
Prasher, D. C., Eckenrode, V. K., Ward, W. W., Prendergast, F. G. & Cormier, M. J. Primary structure of the Aequorea victoria green-fluorescent protein. Gene 111, 229–233 (1992).
Chudakov, D. M., Matz, M. V., Lukyanov, S. & Lukyanov, K. A. Fluorescent proteins and their applications in imaging living cells and tissues. Physiol. Rev. 90, 1103–1163 (2010).
Kremers, G. J., Gilbert, S. G., Cranfill, P. J., Davidson, M. W. & Piston, D. W. Fluorescent proteins at a glance. J. Cell Sci. 124, 157–160 (2011).
Weber, K., Bartsch, U., Stocking, C. & Fehse, B. A multicolor panel of novel lentiviral “gene ontology” (LeGO) vectors for functional gene analysis. Mol. Ther. 16, 698–706 (2008).
Weber, K., Mock, U., Petrowitz, B., Bartsch, U. & Fehse, B. Lentiviral gene ontology (LeGO) vectors equipped with novel drug-selectable fluorescent proteins: new building blocks for cell marking and multi-gene analysis. Gene Ther. 17, 511–520 (2010).
Weber, K., Thomaschewski, M., Benten, D. & Fehse, B. RGB marking with lentiviral vectors for multicolor clonal cell tracking. Nat. Protoc. 7, 839–849 (2012). This article provides a highly detailed protocol for the use of red–green–blue labelling using LeGO vectors.
Gomez-Nicola, D., Riecken, K., Fehse, B. & Perry, V. H. In-vivo RGB marking and multicolour single-cell tracking in the adult brain. Sci. Rep. 4, 7520 (2014).
Mohme, M. et al. Optical barcoding for single-clone tracking to study tumor heterogeneity. Mol. Ther. 25, 621–633 (2017). This study provides one of the first demonstrations highlighting the power of LeGO vectors to study cancer heterogeneity.
Shembrey, C. et al. Longitudinal monitoring of intra-tumoural heterogeneity using optical barcoding of patient-derived colorectal tumour models. Cancers https://doi.org/10.3390/cancers14030581 (2022).
Marusyk, A. & Polyak, K. Tumor heterogeneity: causes and consequences. Biochim. Biophys. Acta 1805, 105–117 (2010).
Singh, S. K. et al. Identification of human brain tumour initiating cells. Nature 432, 396–401 (2004).
Zhang, M. et al. Identification of tumor-initiating cells in a p53-null mouse model of breast cancer. Cancer Res. 68, 4674–4682 (2008).
Echeverria, G. V. et al. High-resolution clonal mapping of multi-organ metastasis in triple negative breast cancer. Nat. Commun. 9, 5079 (2018).
Echeverria, G. V. et al. Resistance to neoadjuvant chemotherapy in triple-negative breast cancer mediated by a reversible drug-tolerant state. Sci. Transl. Med. https://doi.org/10.1126/scitranslmed.aav0936 (2019).
Lan, X. et al. Fate mapping of human glioblastoma reveals an invariant stem cell hierarchy. Nature 549, 227–232 (2017).
van der Heijden, M. et al. Spatiotemporal regulation of clonogenicity in colorectal cancer xenografts. Proc. Natl Acad. Sci. USA 116, 6140–6145 (2019).
Nguyen, L. V. et al. Barcoding reveals complex clonal dynamics of de novo transformed human mammary cells. Nature 528, 267–271 (2015).
Maire, C. L. et al. Glioma escape signature and clonal development under immune pressure. J. Clin. Invest. 130, 5257–5271 (2020).
Baldwin, L. A. et al. DNA barcoding reveals ongoing immunoediting of clonal cancer populations during metastatic progression and in response to immunotherapy. Preprint at bioRxiv https://doi.org/10.1101/2021.01.11.426174 (2021).
Wagenblast, E. et al. A model of breast cancer heterogeneity reveals vascular mimicry as a driver of metastasis. Nature 520, 358–362 (2015).
Iacobuzio-Donahue, C. A. et al. Cancer biology as revealed by the research autopsy. Nat. Rev. Cancer 19, 686–697 (2019).
Paget, S. The distribution of secondary growths in cancer of the breast. Lancet 133, 571–573 (1889).
Lin, D. S. et al. DiSNE movie visualization and assessment of clonal kinetics reveal multiple trajectories of dendritic cell development. Cell Rep. 22, 2557–2566 (2018).
Tian, L. et al. Clonal multi-omics reveals Bcor as a negative regulator of emergency dendritic cell development. Immunity 54, 1338–1351 e1339 (2021).
Maetzig, T. et al. Lentiviral fluorescent genetic barcoding for multiplex fate tracking of leukemic cells. Mol. Ther. Methods Clin. Dev. 6, 54–65 (2017).
Ben-David, U. et al. Genetic and transcriptional evolution alters cancer cell line drug response. Nature 560, 325–330 (2018).
Birsoy, K. et al. Metabolic determinants of cancer cell sensitivity to glucose limitation and biguanides. Nature 508, 108–112 (2014).
Li, H. et al. The landscape of cancer cell line metabolism. Nat. Med. 25, 850–860 (2019).
Garcia-Bermudez, J. et al. Squalene accumulation in cholesterol auxotrophic lymphomas prevents oxidative cell death. Nature 567, 118–122 (2019).
Akimov, Y., Bulanova, D., Timonen, S., Wennerberg, K. & Aittokallio, T. Improved detection of differentially represented DNA barcodes for high-throughput clonal phenomics. Mol. Syst. Biol. 16, e9195 (2020).
Walens, A. et al. Adaptation and selection shape clonal evolution of tumors during residual disease and recurrence. Nat. Commun. 11, 5017 (2020).
Nolan-Stevaux, O. et al. Measurement of cancer cell growth heterogeneity through lentiviral barcoding identifies clonal dominance as a characteristic of in vivo tumor engraftment. PLoS ONE https://doi.org/10.1371/journal.pone.0067316 (2013).
Rehman, S. K. et al. Colorectal cancer cells enter a diapause-like DTP state to survive chemotherapy. Cell 184, 226–242 e221 (2021).
Milo, I. et al. The immune system profoundly restricts intratumor genetic heterogeneity. Sci. Immunol. https://doi.org/10.1126/sciimmunol.aat1435 (2018).
Jin, X. et al. A metastasis map of human cancer cell lines. Nature 588, 331–336 (2020). This study uses the multiplexing capability of genetic barcoding to generate a ‘metastasis map’ for 500 cancer cell lines.
Alemany, A., Florescu, M., Baron, C. S., Peterson-Maduro, J. & van Oudenaarden, A. Whole-organism clone tracing using single-cell sequencing. Nature 556, 108–112 (2018).
McKenna, A. et al. Whole-organism lineage tracing by combinatorial and cumulative genome editing. Science 353, aaf7907 (2016).
Frieda, K. L. et al. Synthetic recording and in situ readout of lineage information in single cells. Nature 541, 107–111 (2017).
Raj, B. et al. Simultaneous single-cell profiling of lineages and cell types in the vertebrate brain. Nat. Biotechnol. 36, 442–450 (2018).
Simeonov, K. P. et al. Single-cell lineage tracing of metastatic cancer reveals selection of hybrid EMT states. Cancer Cell 39, 1150–1162 e1159 (2021).
Gambera, S. et al. Clonal dynamics in osteosarcoma defined by RGB marking. Nat. Commun. 9, 3994 (2018).
Lamprecht, S. et al. Multicolor lineage tracing reveals clonal architecture and dynamics in colon cancer. Nat. Commun. 8, 1406 (2017).
Tiede, S. et al. Multi-color clonal tracking reveals intra-stage proliferative heterogeneity during mammary tumor progression. Oncogene 40, 12–27 (2021).
Kalhor, R., Mali, P. & Church, G. M. Rapidly evolving homing CRISPR barcodes. Nat. Methods 14, 195–200 (2017).
Zhang, W. et al. The bone microenvironment invigorates metastatic seeds for further dissemination. Cell 184, 2471–2486 e2420 (2021).
Quinn, J. J. et al. Single-cell lineages reveal the rates, routes, and drivers of metastasis in cancer xenografts. Science https://doi.org/10.1126/science.abc1944 (2021).
Pei, W. et al. Using Cre-recombinase-driven Polylox barcoding for in vivo fate mapping in mice. Nat. Protoc. 14, 1820–1840 (2019).
Weber, T. S. et al. Site-specific recombinatorics: in situ cellular barcoding with the Cre Lox system. BMC Syst. Biol. 10, 43 (2016).
Sun, J. et al. Clonal dynamics of native haematopoiesis. Nature 514, 322–327 (2014).
Bowling, S. et al. An engineered CRISPR-Cas9 mouse line for simultaneous readout of lineage histories and gene expression profiles in single cells. Cell 181, 1410–1422 e1427 (2020).
Yang, D. et al. Lineage tracing reveals the phylodynamics, plasticity, and paths of tumor evolution. Cell https://doi.org/10.1016/j.cell.2022.04.015 (2022).
Yu, C. et al. High-throughput identification of genotype-specific cancer vulnerabilities in mixtures of barcoded tumor cell lines. Nat. Biotechnol. 34, 419–423 (2016).
Patwardhan, G. A. et al. Treatment scheduling effects on the evolution of drug resistance in heterogeneous cancer cell populations. NPJ Breast Cancer 7, 60 (2021).
Gonzalez Rajal, A. et al. A non-genetic, cell cycle-dependent mechanism of platinum resistance in lung adenocarcinoma. Elife https://doi.org/10.7554/eLife.65234 (2021).
Hata, A. N. et al. Tumor cells can follow distinct evolutionary paths to become resistant to epidermal growth factor receptor inhibition. Nat. Med. 22, 262–269 (2016).
Bhang, H. E. et al. Studying clonal dynamics in response to cancer therapy using high-complexity barcoding. Nat. Med. 21, 440–448 (2015).
Hinohara, K. et al. KDM5 histone demethylase activity links cellular transcriptomic heterogeneity to therapeutic resistance. Cancer Cell 34, 939–953 e939 (2018).
Marine, J. C., Dawson, S. J. & Dawson, M. A. Non-genetic mechanisms of therapeutic resistance in cancer. Nat. Rev. Cancer 20, 743–756 (2020).
Kaufman, T. et al. Visual barcodes for clonal-multiplexing of live microscopy-based assays. Nat. Commun. 13, 2725 (2022). In this study, fluorescent proteins targeted to specific locations are used to increase the multiplexing potential of optical barcoding.
Roh, V. et al. Cellular barcoding identifies clonal substitution as a hallmark of local recurrence in a surgical model of head and neck squamous cell carcinoma. Cell Rep. 25, 2208–2222 e2207 (2018).
Maetzig, T., Morgan, M. & Schambach, A. Fluorescent genetic barcoding for cellular multiplex analyses. Exp. Hematol. 67, 10–17 (2018).
Askary, A. et al. In situ readout of DNA barcodes and single base edits facilitated by in vitro transcription. Nat. Biotechnol. 38, 66–75 (2020).
Eyler, C. E. et al. Single-cell lineage analysis reveals genetic and epigenetic interplay in glioblastoma drug resistance. Genome Biol. 21, 174 (2020).
Gruner, B. M. et al. An in vivo multiplexed small-molecule screening platform. Nat. Methods 13, 883–889 (2016).
Al’Khafaji, A. M., Deatherage, D. & Brock, A. Control of lineage-specific gene expression by functionalized gRNA barcodes. ACS Synth. Biol. 7, 2468–2474 (2018).
Umkehrer, C. et al. Isolating live cell clones from barcoded populations using CRISPRa-inducible reporters. Nat. Biotechnol. 39, 174–178 (2021). This article describes the development of a CaTCH high-diversity library, which is used to study the fate of melanoma clones in response to targeted therapy and retrieve the clones of interest.
Fehse, B., Kustikova, O. S., Bubenheim, M. & Baum, C. Pois(s)on–it’s a question of dose. Gene Ther. 11, 879–881 (2004).
Salehi, S. et al. Clonal fitness inferred from time-series modelling of single-cell cancer genomes. Nature 595, 585–590 (2021).
Ludwig, L. S. et al. Lineage tracing in humans enabled by mitochondrial mutations and single-cell genomics. Cell 176, 1325–1339 e1322 (2019).
Tao, L. et al. Retrospective cell lineage reconstruction in humans by using short tandem repeats. Cell Rep. Methods https://doi.org/10.1016/j.crmeth.2021.100054 (2021).
Nam, A. S. et al. Somatic mutations and cell identity linked by genotyping of transcriptomes. Nature 571, 355–360 (2019).
Madhusoodanan, J. Molecular barcodes reveal tumour lineages. Nature 603, 752–754 (2022).
Rodriguez-Fraticelli, A. & Morris, S. A. In preprints: the fast-paced field of single-cell lineage tracing. Development https://doi.org/10.1242/dev.200877 (2022).
Acknowledgements
The authors acknowledge F. El-Saafin and S. Gadipally for their constructive feedback, and T. Weber for useful discussions. A.S. is supported by the Melbourne Research Scholarship. D.M. is supported by Cancer Council Victoria, the Love Your Sister Foundation, the NBCF (Investigator Initiated Research Scheme grant IIRS-19-082), Susan G. Komen and Cancer Australia (CCR19606878), the Victorian Cancer Agency Mid-Career Research Fellowship (MCRF21011) and the Australian National Health and Medical Research Council (Grant 2012196). S.H.N. is supported by the Australian National Health and Medical Research Council (Grants 1062820, 1100033, 1101378, 1124812 and 1145184). The authors and the Olivia Newton-John Cancer Research Institute gratefully acknowledge the generous support of the Love Your Sister Foundation and the contribution of the Victorian Government acting through the Victorian Cancer Agency. The contents of the published material are solely the responsibility of the individual authors and do not reflect the views of Cancer Australia or other funding agencies.
Author information
Authors and Affiliations
Contributions
D.M. and A.S. researched data for the article. All authors contributed substantially to discussion of the content. D.M. and A.S. wrote the article. All authors reviewed and/or edited the manuscript before submission.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Reviews Cancer thanks Louis Vermeulen and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Glossary
- Clones
-
Subpopulations of cells derived from a common progenitor and harbouring genomic similitude. By extension, in cellular barcoding studies, ‘clone’ refers to cells harbouring the same barcode, as these cells originate from a unique barcoded cell.
- Fitness
-
The ability of individual clones to survive and proliferate in specific conditions.
- Diapause
-
A reversible cellular state originally observed as a survival mechanism of embryos pausing their development when environmental conditions are unfavourable.
- Minimal residual disease
-
Cancer cells detected in a low quantity after treatment.
- Spectral flow cytometry
-
A type of flow cytometry that enables the full spectrum of fluorescence to be recorded with multiple detectors, as opposed to conventional flow cytometry, which uses a single detector.
- Serial dilution experiments
-
Experiments consisting of successive dilution of a cell suspension to select decreasing amounts of cells or to isolate single cells.
- Multiplicity of infection
-
(MOI). The ratio of the number of viral particles to the number of targeted cells.
Rights and permissions
Springer Nature or its licensor holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Serrano, A., Berthelet, J., Naik, S.H. et al. Mastering the use of cellular barcoding to explore cancer heterogeneity. Nat Rev Cancer 22, 609–624 (2022). https://doi.org/10.1038/s41568-022-00500-2
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41568-022-00500-2
This article is cited by
-
Experimental and spontaneous metastasis assays can result in divergence in clonal architecture
Communications Biology (2023)
-
Using picoliter droplet deposition to track clonal competition in adherent and organoid cancer cell cultures
Scientific Reports (2023)
-
Dormant cancer cells: programmed quiescence, senescence, or both?
Cancer and Metastasis Reviews (2023)