Abstract
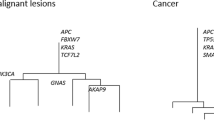
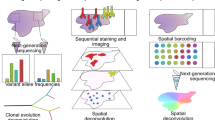
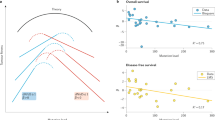
The observation and analysis of intra-tumour heterogeneity (ITH), particularly in genomic studies, has advanced our understanding of the evolutionary forces that shape cancer growth and development. However, only a subset of the variation observed in a single tumour will have an impact on cancer evolution, highlighting the need to distinguish between functional and non-functional ITH. Emerging studies highlight a role for the cancer epigenome, transcriptome and immune microenvironment in functional ITH. Here, we consider the importance of both genetic and non-genetic ITH and their role in tumour evolution, and present the rationale for a broad research focus beyond the cancer genome. Systems-biology analytical approaches will be necessary to outline the scale and importance of functional ITH. By allowing a deeper understanding of tumour evolution this will, in time, encourage development of novel therapies and improve outcomes for patients.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Darwin, C. On the Origin of Species by Means of Natural Selection, or the Preservation of Favored Races in the Struggle for Life (J. Murray, 1859).
Nowell, P. C. The clonal evolution of tumor cell populations. Science 194, 23–28 (1976).
Greaves, M. & Maley, C. C. Clonal evolution in cancer. Nature 481, 306–313 (2012).
Williams, M. J., Werner, B., Graham, T. A. & Sottoriva, A. Functional versus non-functional intratumor heterogeneity in cancer. Mol. Cell. Oncol. 3, e1162897 (2016).
Turajlic, S., Sottoriva, A., Graham, T. & Swanton, C. Resolving genetic heterogeneity in cancer. Nat. Rev. Genet. 20, 404–416 (2019).
Marusyk, A., Janiszewska, M. & Polyak, K. Intratumor heterogeneity: the Rosetta Stone of therapy resistance. Cancer Cell 37, 471–484 (2020).
McGranahan, N., Burrell, R. A., Endesfelder, D., Novelli, M. R. & Swanton, C. Cancer chromosomal instability: therapeutic and diagnostic challenges. EMBO Rep. 13, 528–538 (2012).
Alexandrov, L. B. et al. Clock-like mutational processes in human somatic cells. Nat. Genet. 47, 1402–1407 (2015).
Coons, S. W., Johnson, P. C. & Shapiro, J. R. Cytogenetic and flow cytometry DNA analysis of regional heterogeneity in a low grade human glioma. Cancer Res. 55, 1569–1577 (1995).
Teixeira, M. R., Pandis, N., Bardi, G., Andersen, J. A. & Heim, S. Karyotypic comparisons of multiple tumorous and macroscopically normal surrounding tissue samples from patients with breast cancer. Cancer Res. 56, 855–859 (1996).
Navin, N. et al. Inferring tumor progression from genomic heterogeneity. Genome Res. 20, 68–80 (2010).
Gerlinger, M. et al. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N. Engl. J. Med. 366, 883–892 (2012).
Ohta, T. Slightly deleterious mutant substitutions in evolution. Nature 246, 96–98 (1973).
Ling, S. et al. Extremely high genetic diversity in a single tumor points to prevalence of non-Darwinian cell evolution. Proc. Natl. Acad. Sci. 112, E6496–E6505 (2015).
Sottoriva, A. et al. A big bang model of human colorectal tumor growth. Nat. Genet. 47, 209–216 (2015).
Williams, M. J., Werner, B., Barnes, C. P., Graham, T. A. & Sottoriva, A. Identification of neutral tumor evolution across cancer types. Nat. Genet. 48, 238–244 (2016). This study demonstrates that a subset of tumours evolve without clear evidence of subclonal selection.
Sun, R. et al. Between-region genetic divergence reflects the mode and tempo of tumor evolution. Nat. Genet. 49, 1015–1024 (2017).
Cross, W. et al. The evolutionary landscape of colorectal tumorigenesis. Nat. Ecol. Evol. 2, 1661–1672 (2018).
Baca, S. C. et al. Punctuated evolution of prostate cancer genomes. Cell 153, 666–677 (2013).
Gao, R. et al. Punctuated copy number evolution and clonal stasis in triple-negative breast cancer. Nat. Genet. 48, 1119–1130 (2016).
Notta, F. et al. A renewed model of pancreatic cancer evolution based on genomic rearrangement patterns. Nature 538, 378–382 (2016).
Field, M. G. et al. Punctuated evolution of canonical genomic aberrations in uveal melanoma. Nat. Commun. 9, 116 (2018).
Reiter, J. G. et al. An analysis of genetic heterogeneity in untreated cancers. Nat. Rev. Cancer 19, 639–650 (2019).
Papaemmanuil, E. et al. Clinical and biological implications of driver mutations in myelodysplastic syndromes. Blood 122, 3616–3627 quiz 3699 (2013).
Brastianos, P. K. et al. Genomic characterization of brain metastases reveals branched evolution and potential therapeutic targets. Cancer Discov. 5, 1164–1177 (2015).
Landau, D. A. et al. Mutations driving CLL and their evolution in progression and relapse. Nature 526, 525–530 (2015).
Yates, L. R. et al. Subclonal diversification of primary breast cancer revealed by multiregion sequencing. Nat. Med. 21, 751–759 (2015).
Gerstung, M. et al. The evolutionary history of 2,658 cancers. Nature 578, 122–128 (2020). This study, part of the PCAWG, provides an overview of evolutionary patterns across cancer types, identifying different driver events that typically occur early or late in cancer.
Nik-Zainal, S. et al. The life history of 21 breast cancers. Cell 149, 994–1007 (2012).
Kessler, D. A. & Levine, H. Large population solution of the stochastic Luria–Delbrück evolution model. Proc. Natl Acad. Sci. USA 110, 11682–11687 (2013).
Caravagna, G. et al. Subclonal reconstruction of tumors by using machine learning and population genetics. Nat. Genet. 52, 898–907 (2020).
Litchfield, K. et al. Representative sequencing: unbiased sampling of solid tumor tissue. Cell Rep. 31, 107550 (2020).
Durinck, S. et al. Temporal dissection of tumorigenesis in primary cancers. Cancer Discov. 1, 137–143 (2011).
Martincorena, I. et al. High burden and pervasive positive selection of somatic mutations in normal human skin. Science 348, 880–886 (2015).
Martincorena, I. et al. Universal patterns of selection in cancer and somatic tissues. Cell 171, 1029–1041.e21 (2017).
López, S. et al. Interplay between whole-genome doubling and the accumulation of deleterious alterations in cancer evolution. Nat. Genet. 52, 283–293 (2020).
McGranahan, N. & Swanton, C. Clonal heterogeneity and tumor evolution: past, present, and the future. Cell 168, 613–628 (2017).
Tarabichi, M. et al. Neutral tumor evolution? Nat. Genet. 50, 1630–1633 (2018).
Nam, A. S., Chaligne, R. & Landau, D. A. Integrating genetic and non-genetic determinants of cancer evolution by single-cell multi-omics. Nat. Rev. Genet. 22, 3–18 (2020).
Laks, E. et al. Clonal decomposition and DNA replication states defined by scaled single-cell genome sequencing. Cell 179, 1207–1221.e22 (2019).
Cross, W. et al. Stabilising selection causes grossly altered but stable karyotypes in metastatic colorectal cancer. bioRxiv https://doi.org/10.1101/2020.03.26.007138 (2020).
Turajlic, S. et al. Tracking cancer evolution reveals constrained routes to metastases: TRACERx renal. Cell 173, 581–594.e12 (2018).
Gundem, G. et al. The evolutionary history of lethal metastatic prostate cancer. Nature 520, 353–357 (2015).
Suzuki, H. et al. Mutational landscape and clonal architecture in grade II and III gliomas. Nat. Genet. 47, 458–468 (2015).
Jamal-Hanjani, M. et al. Tracking the evolution of non-small-cell lung cancer. N. Engl. J. Med. 376, 2109–2121 (2017).
Watkins, T. B. K. et al. Pervasive chromosomal instability and karyotype order in tumour evolution. Nature 587, 126–132 (2020). This multi-region, pan-cancer analysis of tumour karyotype uncovers parallel evolution of events within different subclones in one-third of tumours sampled, and identifies the important role of chromosomal instability in generating subclonal diversity in cancer.
Bolhaqueiro, A. C. F. et al. Ongoing chromosomal instability and karyotype evolution in human colorectal cancer organoids. Nat. Genet. 51, 824–834 (2019).
Flavahan, W. A., Gaskell, E. & Bernstein, B. E. Epigenetic plasticity and the hallmarks of cancer. Science 357, eaal2380 (2017).
Clarke, T. L. et al. Histone lysine methylation dynamics control EGFR DNA copy-number amplification. Cancer Discov. 10, 306–325 (2020).
Chen, H. et al. A pan-cancer analysis of enhancer expression in nearly 9000 patient samples. Cell 173, 386–399.e12 (2018).
Gröbner, S. N. et al. The landscape of genomic alterations across childhood cancers. Nature 555, 321–327 (2018).
Kahles, A. et al. Comprehensive analysis of alternative splicing across tumors from 8,705 patients. Cancer Cell 34, 211–224.e6 (2018).
Shiraishi, Y. et al. A comprehensive characterization of cis-acting splicing-associated variants in human cancer. Genome Res. 28, 1111–1125 (2018).
Demircioğlu, D. et al. A pan-cancer transcriptome analysis reveals pervasive regulation through alternative promoters. Cell 178, 1465–1477.e17 (2019).
Calabrese, C. et al. Genomic basis for RNA alterations in cancer. Nature 578, 129–136 (2020). This pan-cancer study of paired whole genomes and transcriptomes illustrates the variety of transcriptomic alterations in cancer, and underlines the influence of copy number events and non-coding mutations on gene expression.
Zhang, Y. et al. High-coverage whole-genome analysis of 1220 cancers reveals hundreds of genes deregulated by rearrangement-mediated cis-regulatory alterations. Nat. Commun. 11, 736 (2020).
Baysal, B. E., Sharma, S., Hashemikhabir, S. & Janga, S. C. RNA editing in pathogenesis of cancer. Cancer Res. 77, 3733–3739 (2017).
Chen, L. et al. Recoding RNA editing of AZIN1 predisposes to hepatocellular carcinoma. Nat. Med. 19, 209–216 (2013).
Bailey, M. H. et al. Comprehensive characterization of cancer driver genes and mutations. Cell 173, 371–385.e18 (2018).
Sondka, Z. et al. The COSMIC Cancer Gene Census: describing genetic dysfunction across all human cancers. Nat. Rev. Cancer 18, 696–705 (2018).
Carter, S. L., Eklund, A. C., Kohane, I. S., Harris, L. N. & Szallasi, Z. A signature of chromosomal instability inferred from gene expression profiles predicts clinical outcome in multiple human cancers. Nat. Genet. 38, 1043–1048 (2006).
Biswas, D. et al. A clonal expression biomarker associates with lung cancer mortality. Nat. Med. 25, 1540–1548 (2019).
Salami, S. S., Tomlins, S. A. & Palapattu, G. S. Transcriptomic heterogeneity in multifocal prostate cancer. JCI Insight 3, e123468 (2018).
Thomsen, M. B. H. et al. Comprehensive multiregional analysis of molecular heterogeneity in bladder cancer. Sci. Rep. 7, 11702 (2017).
Aran, D., Sirota, M. & Butte, A. J. Systematic pan-cancer analysis of tumour purity. Nat. Commun. 6, 8971 (2015).
Wang, F. et al. Integrated transcriptomic–genomic tool Texomer profiles cancer tissues. Nat. Methods 16, 401–404 (2019).
Teschendorff, A. E. & Enver, T. Single-cell entropy for accurate estimation of differentiation potency from a cell’s transcriptome. Nat. Commun. 8, 15599 (2017).
Marjanovic, N. D. et al. Emergence of a high-plasticity cell state during lung cancer evolution. Cancer Cell 38, 229–246 (2020). This study of a mouse model of lung adenocarcinoma illustrates that highly plastic stem-like cells with diverse transcriptional states drive resistance to therapy and poor clinical outcome.
Lafave, L. M. et al. Epigenomic state transitions characterize tumor progression in mouse lung adenocarcinoma. Cancer Cell 38, 212–228.e13 (2020).
Izzo, F. et al. DNA methylation disruption reshapes the hematopoietic differentiation landscape. Nat. Genet. 52, 378–387 (2020).
Raphael, B. J. Integrated genomic characterization of pancreatic ductal adenocarcinoma. Cancer Cell 32, 185–203.e13 (2017).
McDonald, O. G. et al. Epigenomic reprogramming during pancreatic cancer progression links anabolic glucose metabolism to distant metastasis. Nat. Genet. 49, 367–376 (2017). This study of pancreatic cancer is an example of the important potential role of non-genetic variation in cancer evolution.
Pastore, A. et al. Corrupted coordination of epigenetic modifications leads to diverging chromatin states and transcriptional heterogeneity in CLL. Nat. Commun. 10, 1874 (2019).
Hua, X. et al. Genetic and epigenetic intratumor heterogeneity impacts prognosis of lung adenocarcinoma. Nat. Commun. 11, 2459 (2020).
Zhu, B. et al. The genomic and epigenomic evolutionary history of papillary renal cell carcinomas. Nat. Commun. 11, 3096 (2020).
Mertins, P. et al. Proteogenomics connects somatic mutations to signalling in breast cancer. Nature 534, 55–62 (2016).
Zhang, H. et al. Integrated proteogenomic characterization of human high-grade serous ovarian cancer. Cell 166, 755–765 (2016).
Mun, D.-G. et al. Proteogenomic characterization of human early-onset gastric cancer. Cancer Cell 35, 111–124.e10 (2019).
Chen, Y. J. et al. Proteogenomics of non-smoking lung cancer in east Asia delineates molecular signatures of pathogenesis and progression. Cell 182, 226–244.e17 (2020).
Gillette, M. A. et al. Proteogenomic characterization reveals therapeutic vulnerabilities in lung adenocarcinoma. Cell 182, 200–225.e35 (2020).
Xu, J.-Y. et al. Integrative proteomic characterization of human lung adenocarcinoma. Cell 182, 245–261.e17 (2020).
Hanahan, D. & Weinberg, R. A. Hallmarks of cancer: the next generation. Cell 144, 646–674 (2011).
Marty, R. et al. MHC-I genotype restricts the oncogenic mutational landscape. Cell 171, 1272–1283.e15 (2017).
Marty Pyke, R. et al. Evolutionary pressure against MHC class II binding cancer mutations. Cell 175, 416–428.e13 (2018).
DuPage, M., Mazumdar, C., Schmidt, L. M., Cheung, A. F. & Jacks, T. Expression of tumour-specific antigens underlies cancer immunoediting. Nature 482, 405–409 (2012).
Rooney, M. S., Shukla, S. A., Wu, C. J., Getz, G. & Hacohen, N. Molecular and genetic properties of tumors associated with local immune cytolytic activity. Cell 160, 48–61 (2015).
McGranahan, N. et al. Allele-specific HLA loss and immune escape in lung cancer evolution. Cell 171, 1259–1271.e11 (2017).
Anagnostou, V. et al. Evolution of neoantigen landscape during immune checkpoint blockade in non-small cell lung cancer. Cancer Discov. 7, 264–276 (2017).
Rosenthal, R. et al. Neoantigen-directed immune escape in lung cancer evolution. Nature 567, 479–485 (2019). This work highlights the role of immune editing in shaping early cancer evolution by negative selection, as well as the diversity of mechanisms of immune evasion.
Jiménez-Sánchez, A. et al. Unraveling tumor–immune heterogeneity in advanced ovarian cancer uncovers immunogenic effect of chemotherapy. Nat. Genet. 52, 582–593 (2020).
Davoli, T., Uno, H., Wooten, E. C. & Elledge, S. J. Tumor aneuploidy correlates with markers of immune evasion and with reduced response to immunotherapy. Science 355, eaaf8399 (2017).
McGranahan, N. et al. Clonal neoantigens elicit T cell immunoreactivity and sensitivity to immune checkpoint blockade. Science 351, 1463–1470 (2016).
Joshi, K. et al. Spatial heterogeneity of the T cell receptor repertoire reflects the mutational landscape in lung cancer. Nat. Med. 25, 1549–1559 (2019).
Roh, W. et al. Integrated molecular analysis of tumor biopsies on sequential CTLA-4 and PD-1 blockade reveals markers of response and resistance. Sci. Transl. Med. 9, eaah3560 (2017).
Tumeh, P. C. et al. PD-1 blockade induces responses by inhibiting adaptive immune resistance. Nature 515, 568–571 (2014).
Snyder, A. et al. Contribution of systemic and somatic factors to clinical response and resistance to PD-L1 blockade in urothelial cancer: an exploratory multi-omic analysis. PLOS Med. 14, e1002309 (2017).
Wolf, Y. et al. UVB-induced tumor heterogeneity diminishes immune response in melanoma. Cell 179, 219–235.e21 (2019). In this study, a mouse model of melanoma illustrates that increased clonal diversity of a developing tumour is associated with evasion of the anticancer immune response.
Gejman, R. S. et al. Rejection of immunogenic tumor clones is limited by clonal fraction. eLife 7, e41090 (2018).
Maire, C. L. et al. Glioma escape signature and clonal development under immune pressure. J. Clin. Invest. 130, 5257–5271 (2020).
Zhang, A. W. et al. Interfaces of malignant and immunologic clonal dynamics in ovarian cancer. Cell 173, 1755–1769.e22 (2018).
Angelova, M. et al. Evolution of metastases in space and time under immune selection. Cell 175, 751–765.e16 (2018).
Pennycuick, A. et al. Immune surveillance in clinical regression of pre-invasive squamous cell lung cancer. Cancer Discov. 10, 1489–1499 (2020).
Zapata, L. et al. Negative selection in tumor genome evolution acts on essential cellular functions and the immunopeptidome. Genome Biol. 19, 67 (2018).
Eynden, J. Van Den, Jiménez-sánchez, A., Miller, M. L. & Larsson, E. Lack of detectable neoantigen depletion signals in the untreated cancer genome. Nat. Genet. 51, 1741–1748 (2019).
Zapata, L. et al. dN/dS dynamics quantify tumour immunogenicity and predict response to immunotherapy. bioRxiv https://doi.org/10.1101/2020.07.21.215038 (2020).
Ghorani, E. et al. The T cell differentiation landscape is shaped by tumour mutations in lung cancer. Nat. Cancer 1, 546–561 (2020).
Keren, L. et al. A structured tumor-immune microenvironment in triple negative breast cancer revealed by multiplexed ion beam imaging. Cell 174, 1373–1387.e19 (2018).
Abduljabbar, K. et al. Geospatial immune variability illuminates differential evolution of lung adenocarcinoma. Nat. Med. 26, 1054–1062 (2020).
Wang, M. et al. Role of tumor microenvironment in tumorigenesis. J. Cancer 8, 761–773 (2017).
Korenchan, D. E. & Flavell, R. R. Spatiotemporal pH heterogeneity as a promoter of cancer progression and therapeutic resistance. Cancers 11, 1026 (2019).
Lloyd, M. C. et al. Darwinian dynamics of intratumoral heterogeneity: not solely random mutations but also variable environmental selection forces. Cancer Res. 76, 3136–3144 (2016).
Hoefflin, R. et al. Spatial niche formation but not malignant progression is a driving force for intratumoural heterogeneity. Nat. Commun. 7, 11845 (2016).
Lujambio, A. et al. Non-cell-autonomous tumor suppression by p53. Cell 153, 449–460 (2013).
Peinado, H. et al. Pre-metastatic niches: organ-specific homes for metastases. Nat. Rev. Cancer 17, 302–317 (2017).
Montagner, M. et al. Crosstalk with lung epithelial cells regulates Sfrp2-mediated latency in breast cancer dissemination. Nat. Cell Biol. 22, 289–296 (2020).
Marusyk, A. et al. Non-cell-autonomous driving of tumour growth supports sub-clonal heterogeneity. Nature 514, 54–58 (2014).
Schürch, C. M. et al. Coordinated cellular neighborhoods orchestrate antitumoral immunity at the colorectal cancer invasive front. Cell 182, 1341–1359.e19 (2020).
Wagner, J. et al. A single-cell atlas of the tumor and immune ecosystem of human breast cancer. Cell 177, 1330–1345.e18 (2019).
Ali, H. R. et al. Imaging mass cytometry and multiplatform genomics define the phenogenomic landscape of breast cancer. Nat. Cancer 1, 163–175 (2020).
Jackson, H. W. et al. The single-cell pathology landscape of breast cancer. Nature 578, 615–620 (2020).
Birkbak, N. J. & McGranahan, N. Cancer genome evolutionary trajectories in metastasis. Cancer Cell 37, 8–19 (2020).
Priestley, P. et al. Pan-cancer whole-genome analyses of metastatic solid tumours. Nature 575, 210–216 (2019).
Yates, L. R. et al. Genomic evolution of breast cancer metastasis and relapse. Cancer Cell 32, 169–184.e7 (2017).
Hu, Z., Li, Z., Ma, Z. & Curtis, C. Multi-cancer analysis of clonality and the timing of systemic spread in paired primary tumors and metastases. Nat. Genet. 52, 701–708 (2020).
Reiter, J. G. et al. Lymph node metastases develop through a wider evolutionary bottleneck than distant metastases. Nat. Genet. 52, 692–700 (2020).
El-Kebir, M., Satas, G. & Raphael, B. J. Inferring parsimonious migration histories for metastatic cancers. Nat. Genet. 50, 718–726 (2018).
Reiter, J. G. et al. Minimal functional driver gene heterogeneity among untreated metastases. Science 361, 1033–1037 (2018).
Rabbie, R. et al. Multi-site clonality analysis uncovers pervasive heterogeneity across melanoma metastases. Nat. Commun. 11, 4306 (2020).
US National Library of Medicine. ClinicalTrials.gov https://clinicaltrials.gov/ct2/show/NCT03004755 (2019).
Parikh, A. R. et al. Liquid versus tissue biopsy for detecting acquired resistance and tumor heterogeneity in gastrointestinal cancers. Nat. Med. 25, 1415–1421 (2019).
Noorani, A. et al. Genomic evidence supports a clonal diaspora model for metastases of esophageal adenocarcinoma. Nat. Genet. 52, 74–83 (2020).
Lo, H. C. et al. Resistance to natural killer cell immunosurveillance confers a selective advantage to polyclonal metastasis. Nat. Cancer 1, 709–722 (2020).
Landau, D. A. et al. Evolution and impact of subclonal mutations in chronic lymphocytic leukemia. Cell 152, 714–726 (2013).
Andor, N. et al. Pan-cancer analysis of the extent and consequences of intratumor heterogeneity. Nat. Med. 22, 105–113 (2016).
Rye, I. H. et al. Intratumor heterogeneity defines treatment-resistant HER2+ breast tumors. Mol. Oncol. 12, 1838–1855 (2018).
Turajlic, S. et al. Deterministic evolutionary trajectories influence primary tumor growth: TRACERx renal. Cell 173, 595–610.e11 (2018).
Cerrano, M. et al. Prognostic impact of clonal diversity in acute myeloid leukemia (AML) treated with intensive chemotherapy (IC). Blood 134, 2700 (2019).
Iacobuzio-Donahue, C. A., Litchfield, K. & Swanton, C. Intratumor heterogeneity reflects clinical disease course. Nat. Cancer 1, 3–6 (2020).
Martinez, P. et al. Dynamic clonal equilibrium and predetermined cancer risk in Barrett’s oesophagus. Nat. Commun. 7, 12158 (2016).
Wei, S. C., Duffy, C. R. & Allison, J. P. Fundamental mechanisms of immune checkpoint blockade therapy. Cancer Discov. 8, 1069–1086 (2018).
Birkbak, N. J. et al. Paradoxical relationship between chromosomal instability and survival outcome in cancer. Cancer Res. 71, 3447–3452 (2011).
Jamal-Hanjani, M. et al. Extreme chromosomal instability forecasts improved outcome in ER-negative breast cancer: a prospective validation cohort study from the TACT trial. Ann. Oncol. 26, 1340–1346 (2015).
Lawson, A. R. J. et al. Extensive heterogeneity in somatic mutation and selection in the human bladder. Science 370, 75–82 (2020).
Moore, L. et al. The mutational landscape of normal human endometrial epithelium. Nature 580, 640–646 (2020).
Killcoyne, S. et al. Genomic copy number predicts esophageal cancer years before transformation. Nat. Med. 26, 1726–1732 (2020).
Noble, R. & Burley, J. T. When, why and how tumour clonal diversity predicts survival. Evol. Appl. 13, 1558–1568 (2020).
Riaz, N. et al. Tumor and microenvironment evolution during immunotherapy with nivolumab. Cell 171, 934–949.e16 (2017).
Pearson, A. et al. High-level clonal FGFR amplification and response to FGFR inhibition in a translational clinical trial. Cancer Discov. 6, 838–851 (2016).
Russo, M. et al. Adaptive mutability of colorectal cancers in response to targeted therapies. Science 366, 1473–1480 (2019).
Mok, T. S. et al. Osimertinib or platinum-pemetrexed in EGFR T790M-positive lung cancer. N. Engl. J. Med. 376, 629–640 (2017).
Oldrini, B. et al. MGMT genomic rearrangements contribute to chemotherapy resistance in gliomas. Nat. Commun. 11, 3883 (2020).
Bell, C. C. et al. Targeting enhancer switching overcomes non-genetic drug resistance in acute myeloid leukaemia. Nat. Commun. 10, 2723 (2019).
Fong, C. Y. et al. BET inhibitor resistance emerges from leukaemia stem cells. Nature 525, 538–542 (2015).
Hinohara, K. et al. KDM5 histone demethylase activity links cellular transcriptomic heterogeneity to therapeutic resistance. Cancer Cell 34, 939–953.e9 (2018).
McGranahan, N. & Swanton, C. Neoantigen quality, not quantity. Sci. Transl. Med. 11, eaax7918 (2019).
Rambow, F. et al. Toward minimal residual disease-directed therapy in melanoma. Cell 174, 843–855.e19 (2018). This work highlights the role played by stem-like cancer cells in non-genetic mechanisms of resistance to cancer therapy.
Pleasance, E. et al. Pan-cancer analysis of advanced patient tumors reveals interactions between therapy and genomic landscapes. Nat. Cancer 1, 452–468 (2020).
Gao, J. et al. Loss of IFN-γ pathway genes in tumor cells as a mechanism of resistance to anti-CTLA-4 therapy. Cell 167, 397–404.e9 (2016).
Sahin, U. et al. Personalized RNA mutanome vaccines mobilize poly-specific therapeutic immunity against cancer. Nature 547, 222–226 (2017).
Siravegna, G. et al. Clonal evolution and resistance to EGFR blockade in the blood of colorectal cancer patients. Nat. Med. 21, 795–801 (2015).
Xue, Y. et al. An approach to suppress the evolution of resistance in BRAF(V600E)-mutant cancer. Nat. Med. 23, 929–937 (2017).
Gopal, P. et al. Clonal selection confers distinct evolutionary trajectories in BRAF-driven cancers. Nat. Commun. 10, 5143 (2019).
Acar, A. et al. Exploiting evolutionary steering to induce collateral drug sensitivity in cancer. Nat. Commun. 11, 1923 (2020).
Algazi, A. et al. Abstract CT013: SWOG S1320: improved progression-free survival with continuous compared to intermittent dosing with dabrafenib and trametinib in patients with BRAF mutated melanoma. Cancer Res. 80, CT013 LP–CT013 (2020).
Acknowledgements
The authors thank C. Bailey, A. Frankell, E. Colliver, C.-M. Ruiz and J. Demeulemeester for their thoughtful review of the manuscript. N.M is a Sir Henry Dale Fellow, jointly funded by the Wellcome Trust and the Royal Society (Grant Number 211179/Z/18/Z), and also receives funding from Cancer Research UK Lung Cancer Centre of Excellence, Rosetrees, and the NIHR BRC at University College London Hospitals.
Author information
Authors and Affiliations
Contributions
J.R.M.B. and N.M. both researched data for the article and made a substantial contribution to discussion of content, writing, reviewing and editing the article.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests. N.M. has received consultancy fees and has stock options in Achilles Therapeutics. N.M. holds European patents relating to targeting neoantigens (PCT/EP2016/ 059401), identifying patient response to immune checkpoint blockade (PCT/ EP2016/071471), determining HLA LOH (PCT/GB2018/052004), predicting survival rates of patients with cancer (PCT/GB2020/050221). J.R.M.B. declares no competing interests.
Additional information
Peer review information
Nature Reviews Cancer thanks M. Gerstung and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Glossary
- Intra-tumour heterogeneity
-
(ITH). Variation within the same tumour; this can be non-functional, a result of neutral evolution, or functional, leading to selection that shapes ongoing tumour evolution.
- Chromosomal instability
-
(CIN). A defect in which cells can gain, lose or rearrange parts of chromosomes or whole chromosomes during cell division; this is a source of variation in cancer.
- Chromothripsis
-
A mutational process in which large numbers of clustered structural rearrangements occur in single or multiple chromosomes.
- Molecular time
-
An estimate of the timing of an event, from the first cell division following fertilization to a cell division that occurred only recently before sampling.
- Enhancer
-
A short genomic region that influences the expression of another gene in cis.
Rights and permissions
About this article
Cite this article
Black, J.R.M., McGranahan, N. Genetic and non-genetic clonal diversity in cancer evolution. Nat Rev Cancer 21, 379–392 (2021). https://doi.org/10.1038/s41568-021-00336-2
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41568-021-00336-2
This article is cited by
-
Joint analysis of mutational and transcriptional landscapes in human cancer reveals key perturbations during cancer evolution
Genome Biology (2024)
-
Challenges in developing personalized neoantigen cancer vaccines
Nature Reviews Immunology (2024)
-
Characterization of tumor evolution by functional clonality and phylogenetics in hepatocellular carcinoma
Communications Biology (2024)
-
Towards targeting transposable elements for cancer therapy
Nature Reviews Cancer (2024)
-
Single-mitosis dissection of acute and chronic DNA mutagenesis and repair
Nature Genetics (2024)