Abstract
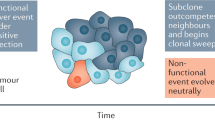
Cancer is a clonal disorder derived from a single ancestor cell and its progenies that are positively selected by acquisition of ‘driver mutations’. However, the evolution of positively selected clones does not necessarily imply the presence of cancer. On the contrary, it has become clear that expansion of these clones in phenotypically normal or non-cancer tissues is commonly seen in association with ageing and/or in response to environmental insults and chronic inflammation. Recent studies have reported expansion of clones harbouring mutations in cancer driver genes in the blood, skin, oesophagus, bronchus, liver, endometrium and bladder, where the expansion could be so extensive that tissues undergo remodelling of an almost entire tissue. The presence of common cancer driver mutations in normal tissues suggests a strong link to cancer development, providing an opportunity to understand early carcinogenic processes. Nevertheless, some driver mutations are unique to normal tissues or have a mutation frequency that is much higher in normal tissue than in cancer, indicating that the respective clones may not necessarily be destined for evolution to cancer but even negatively selected for carcinogenesis depending on the mutated gene. Moreover, tissues that are remodelled by genetically altered clones might define functionalities of aged tissues or modified inflammatory processes. In this Review, we provide an overview of major findings on clonal expansion in phenotypically normal or non-cancer tissues and discuss their biological significance not only in cancer development but also in ageing and inflammatory diseases.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Greenman, C. et al. Patterns of somatic mutation in human cancer genomes. Nature 446, 153–158 (2007).
Greaves, M. & Maley, C. C. Clonal evolution in cancer. Nature 481, 306–313 (2012).
Vogelstein, B. et al. Cancer genome landscapes. Science 339, 1546–1558 (2013).
McGranahan, N. & Swanton, C. Clonal heterogeneity and tumor evolution: past, present, and the future. Cell 168, 613–628 (2017).
Lawrence, M. S. et al. Mutational heterogeneity in cancer and the search for new cancer-associated genes. Nature 499, 214–218 (2013).
Martincorena, I. et al. Universal patterns of selection in cancer and somatic tissues. Cell 171, 1029–1041.e21 (2017).
Bailey, M. H. et al. Comprehensive characterization of cancer driver genes and mutations. Cell 173, 371–385.e18 (2018).
The ICGC/TCGA Pan-Cancer Analysis of Whole Genomes Consortium et al. Pan-cancer analysis of whole genomes. Nature 578, 82–93 (2020).
Alexandrov, L. B. et al. Signatures of mutational processes in human cancer. Nature 500, 415–421 (2013).
Alexandrov, L. B. et al. The repertoire of mutational signatures in human cancer. Nature 578, 94–101 (2020).
Burrell, R. A., McGranahan, N., Bartek, J. & Swanton, C. The causes and consequences of genetic heterogeneity in cancer evolution. Nature 501, 338–345 (2013).
Yachida, S. et al. Distant metastasis occurs late during the genetic evolution of pancreatic cancer. Nature 467, 1114–1117 (2010).
Gerlinger, M. et al. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N. Engl. J. Med. 366, 883–892 (2012).
Turajlic, S. et al. Tracking cancer evolution reveals constrained routes to metastases: TRACERx renal. Cell 173, 581–594.e12 (2018).
Mitchell, T. J. et al. Timing the landmark events in the evolution of clear cell renal cell cancer: TRACERx renal. Cell 173, 611–623.e17 (2018).
Suzuki, H. et al. Mutational landscape and clonal architecture in grade II and III gliomas. Nat. Genet. 47, 458–468 (2015).
Mazor, T. et al. DNA methylation and somatic mutations converge on the cell cycle and define similar evolutionary histories in brain tumors. Cancer Cell 28, 307–317 (2015).
Nikbakht, H. et al. Spatial and temporal homogeneity of driver mutations in diffuse intrinsic pontine glioma. Nat. Commun. 7, 11185 (2016).
Zhang, J. et al. Intratumor heterogeneity in localized lung adenocarcinomas delineated by multiregion sequencing. Science 346, 256–259 (2014).
de Bruin, E. C. et al. Spatial and temporal diversity in genomic instability processes defines lung cancer evolution. Science 346, 251–256 (2014).
Jamal-Hanjani, M. et al. Tracking the evolution of non-small-cell lung cancer. N. Engl. J. Med. 376, 2109–2121 (2017).
Sottoriva, A. et al. A Big Bang model of human colorectal tumor growth. Nat. Genet. 47, 209–216 (2015).
Uchi, R. et al. Integrated multiregional analysis proposing a new model of colorectal cancer evolution. PLoS Genet. 12, e1005778 (2016).
Roerink, S. F. et al. Intra-tumour diversification in colorectal cancer at the single-cell level. Nature 556, 457–462 (2018).
Saito, T. et al. A temporal shift of the evolutionary principle shaping intratumor heterogeneity in colorectal cancer. Nat. Commun. 9, 2884 (2018).
Gundem, G. et al. The evolutionary history of lethal metastatic prostate cancer. Nature 520, 353–357 (2015).
Cooper, C. S. et al. Analysis of the genetic phylogeny of multifocal prostate cancer identifies multiple independent clonal expansions in neoplastic and morphologically normal prostate tissue. Nat. Genet. 47, 367–372 (2015).
Shain, A. H. et al. The genetic evolution of melanoma from precursor lesions. N. Engl. J. Med. 373, 1926–1936 (2015).
Harbst, K. et al. Multiregion whole-exome sequencing uncovers the genetic evolution and mutational heterogeneity of early-stage metastatic melanoma. Cancer Res. 76, 4765–4774 (2016).
Bashashati, A. et al. Distinct evolutionary trajectories of primary high-grade serous ovarian cancers revealed through spatial mutational profiling. J. Pathol. 231, 21–34 (2013).
Eckert, M. A. et al. Genomics of ovarian cancer progression reveals diverse metastatic trajectories including intraepithelial metastasis to the fallopian tube. Cancer Discov. 6, 1342–1351 (2016).
Hao, J. J. et al. Spatial intratumoral heterogeneity and temporal clonal evolution in esophageal squamous cell carcinoma. Nat. Genet. 48, 1500–1507 (2016).
Murugaesu, N. et al. Tracking the genomic evolution of esophageal adenocarcinoma through neoadjuvant chemotherapy. Cancer Discov. 5, 821–831 (2015).
Bolli, N. et al. Heterogeneity of genomic evolution and mutational profiles in multiple myeloma. Nat. Commun. 5, 2997 (2014).
Yates, L. R. et al. Subclonal diversification of primary breast cancer revealed by multiregion sequencing. Nat. Med. 21, 751–759 (2015).
Xue, R. et al. Variable intra-tumor genomic heterogeneity of multiple lesions in patients with hepatocellular carcinoma. Gastroenterology 150, 998–1008 (2016).
Genovese, G. et al. Clonal hematopoiesis and blood-cancer risk inferred from blood DNA sequence. N. Engl. J. Med. 371, 2477–2487 (2014).
Jaiswal, S. et al. Age-related clonal hematopoiesis associated with adverse outcomes. N. Engl. J. Med. 371, 2488–2498 (2014). Together with Genovese et al. (2014), this landmark study describes age-related clonal haematopoiesis having common mutations in myeloid neoplasms, using unbiased sequencing and its relationship not only to haematological malignancy but also to cardiovascular diseases.
Yoshizato, T. et al. Somatic mutations and clonal hematopoiesis in aplastic anemia. N. Engl. J. Med. 373, 35–47 (2015). This study describes clonal haematopoiesis in patients with aplastic anaemia and its link to the clinical leukaemia risk and response to immunosuppressive therapy.
Martincorena, I. et al. High burden and pervasive positive selection of somatic mutations in normal human skin. Science 348, 880–886 (2015). This study shows clonal expansion in sun-exposed skin by targeted sequencing of common drivers in skin cancers, and is the first study demonstrating clonal expansion in normal solid tissues.
Ross-Innes, C. S. et al. Whole-genome sequencing provides new insights into the clonal architecture of Barrett’s esophagus and esophageal adenocarcinoma. Nat. Genet. 47, 1038–1046 (2015).
Stachler, M. D. et al. Paired exome analysis of Barrett’s esophagus and adenocarcinoma. Nat. Genet. 47, 1047–1055 (2015).
Huang, K. K. et al. Genomic and epigenomic profiling of high-risk intestinal metaplasia reveals molecular determinants of progression to gastric cancer. Cancer Cell 33, 137–150.e5 (2018).
Martincorena, I. et al. Somatic mutant clones colonize the human esophagus with age. Science 362, 911–917 (2018).
Yokoyama, A. et al. Age-related remodelling of oesophageal epithelia by mutated cancer drivers. Nature 565, 312–317 (2019). This study shows extensive remodelling of the oesophageal epithelium with ageing, which is substantially accelerated by heavy smoking and alcohol drinking, and comprehensively describes driver mutations and their history.
Kim, S. K. et al. Comprehensive analysis of genetic aberrations linked to tumorigenesis in regenerative nodules of liver cirrhosis. J. Gastroenterol. 54, 628–640 (2019).
Brunner, S. F. et al. Somatic mutations and clonal dynamics in healthy and cirrhotic human liver. Nature 574, 538–542 (2019). This study shows that clonal expansion in cirrhotic liver does not necessarily accompany driver mutations.
Kakiuchi, N. et al. Frequent mutations that converge on the NFKBIZ pathway in ulcerative colitis. Nature 577, 260–265 (2020). This study describes crypt expansion throughout life in the healthy colon, positive selection in ulcerative colitis and negative selection of positively selected clones during carcinogenesis.
Nanki, K. et al. Somatic inflammatory gene mutations in human ulcerative colitis epithelium. Nature 577, 254–259 (2020). This study describes positive selection of clones in ulcerative colitis.
Moore, L. et al. The mutational landscape of normal human endometrial epithelium. Nature 580, 640–646 (2020). This study describes positive selection in endometrial glands in which the majority of glands acquire driver mutations in older people.
Yoshida, K. et al. Tobacco smoking and somatic mutations in human bronchial epithelium. Nature 578, 266–272 (2020). This study describes a mutational landscape of the bronchial epithelium according to past and current history of tobacco smoking. Once remodelled by driver-mutated clones, bronchial epithelium clones can be replaced by mitotically quiescent cells after smoking cessation.
Fearon, E. R., Burke, P. J., Schiffer, C. A., Zehnbauer, B. A. & Vogelstein, B. Differentiation of leukemia cells to polymorphonuclear leukocytes in patients with acute nonlymphocytic leukemia. N. Engl. J. Med. 315, 15–24 (1986).
Fialkow, P. J. et al. Clonal development, stem-cell differentiation, and clinical remissions in acute nonlymphocytic leukemia. N. Engl. J. Med. 317, 468–473 (1987).
Fey, M. F. et al. Clonality and X-inactivation patterns in hematopoietic cell populations detected by the highly informative M27 β DNA probe. Blood 83, 931–938 (1994).
Busque, L. et al. Nonrandom X-inactivation patterns in normal females: lyonization ratios vary with age. Blood 88, 59–65 (1996).
van Kamp, H., Landegent, J. E., Jansen, R. P., Willemze, R. & Fibbe, W. E. Clonal hematopoiesis in patients with acquired aplastic anemia. Blood 78, 3209–3214 (1991).
Taylor, R. W. et al. Mitochondrial DNA mutations in human colonic crypt stem cells. J. Clin. Invest. 112, 1351–1360 (2003).
Greaves, L. C. et al. Mitochondrial DNA mutations are established in human colonic stem cells, and mutated clones expand by crypt fission. Proc. Natl Acad. Sci. USA 103, 714–719 (2006).
Baker, A. M. et al. Quantification of crypt and stem cell evolution in the normal and neoplastic human colon. Cell Rep. 8, 940–947 (2014).
McDonald, S. A. et al. Mechanisms of field cancerization in the human stomach: the expansion and spread of mutated gastric stem cells. Gastroenterology 134, 500–510 (2008).
Gutierrez-Gonzalez, L. et al. The clonal origins of dysplasia from intestinal metaplasia in the human stomach. Gastroenterology 140, 1251–1260.e1–6 (2011).
Gutierrez-Gonzalez, L. et al. Analysis of the clonal architecture of the human small intestinal epithelium establishes a common stem cell for all lineages and reveals a mechanism for the fixation and spread of mutations. J. Pathol. 217, 489–496 (2009).
Fellous, T. G. et al. A methodological approach to tracing cell lineage in human epithelial tissues. Stem Cell 27, 1410–1420 (2009).
Lin, W. R. et al. The histogenesis of regenerative nodules in human liver cirrhosis. Hepatology 51, 1017–1026 (2010).
Cereser, B. et al. Analysis of clonal expansions through the normal and premalignant human breast epithelium reveals the presence of luminal stem cells. J. Pathol. 244, 61–70 (2018).
Gaisa, N. T. et al. The human urothelium consists of multiple clonal units, each maintained by a stem cell. J. Pathol. 225, 163–171 (2011).
Gaisa, N. T. et al. Clonal architecture of human prostatic epithelium in benign and malignant conditions. J. Pathol. 225, 172–180 (2011).
Moad, M. et al. Multipotent basal stem cells, maintained in localized proximal niches, support directed long-ranging epithelial flows in human prostates. Cell Rep. 20, 1609–1622 (2017).
Mitani, K. et al. Somatic mosaicism in chronic myeloid leukemia in remission. Blood 128, 2863–2866 (2016).
Ziegler, A. et al. Sunburn and p53 in the onset of skin cancer. Nature 372, 773–776 (1994).
Nakazawa, H. et al. UV and skin cancer: specific p53 gene mutation in normal skin as a biologically relevant exposure measurement. Proc. Natl Acad. Sci. USA 91, 360–364 (1994).
Jonason, A. S. et al. Frequent clones of p53-mutated keratinocytes in normal human skin. Proc. Natl Acad. Sci. USA 93, 14025–14029 (1996).
Ling, G. et al. Persistent p53 mutations in single cells from normal human skin. Am. J. Pathol. 159, 1247–1253 (2001).
Tian, D. et al. Multifocal accumulation of p53 protein in esophageal carcinoma: evidence for field cancerization. Int. J. Cancer 78, 568–575 (1998).
Katada, C. et al. Alcohol consumption and multiple dysplastic lesions increase risk of squamous cell carcinoma in the esophagus, head, and neck. Gastroenterology 151, 860–869.e7 (2016).
Maley, C. C. et al. The combination of genetic instability and clonal expansion predicts progression to esophageal adenocarcinoma. Cancer Res. 64, 7629–7633 (2004).
Van der Vorst, S., Dekairelle, A. F., Weynand, B., Hamoir, M. & Gala, J. L. Assessment of p53 functional activity in tumor cells and histologically normal mucosa from patients with head and neck squamous cell carcinoma. Head Neck 34, 1542–1550 (2012).
Franklin, W. A. et al. Widely dispersed p53 mutation in respiratory epithelium. A novel mechanism for field carcinogenesis. J. Clin. Invest. 100, 2133–2137 (1997).
Leedham, S. J. et al. Clonality, founder mutations, and field cancerization in human ulcerative colitis-associated neoplasia. Gastroenterology 136, 542–550.e6 (2009).
Galandiuk, S. et al. Field cancerization in the intestinal epithelium of patients with Crohn’s ileocolitis. Gastroenterology 142, 855–864.e8 (2012).
Shimizu, T. et al. Accumulation of somatic mutations in TP53 in gastric epithelium with Helicobacter pylori infection. Gastroenterology 147, 407–417.e3 (2014).
Metzker, M. L. Sequencing technologies — the next generation. Nat. Rev. Genet. 11, 31–46 (2010).
Meyerson, M., Gabriel, S. & Getz, G. Advances in understanding cancer genomes through second-generation sequencing. Nat. Rev. Genet. 11, 685–696 (2010).
Chatterjee, N. & Walker, G. C. Mechanisms of DNA damage, repair, and mutagenesis. Env. Mol. Mutagen. 58, 235–263 (2017).
Welch, J. S. et al. The origin and evolution of mutations in acute myeloid leukemia. Cell 150, 264–278 (2012). This study shows for the first time that normal blood progenitor cells accumulate somatic mutations in an age-dependent manner.
Lodato, M. A. et al. Aging and neurodegeneration are associated with increased mutations in single human neurons. Science 359, 555–559 (2018).
Lee-Six, H. et al. The landscape of somatic mutation in normal colorectal epithelial cells. Nature 574, 532–537 (2019). This study described the mutational signatures and landscape in colon crypts.
Franco, I. et al. Somatic mutagenesis in satellite cells associates with human skeletal muscle aging. Nat. Commun. 9, 800 (2018).
Blokzijl, F. et al. Tissue-specific mutation accumulation in human adult stem cells during life. Nature 538, 260–264 (2016). This study measures the mutational load in normal cells from small intestine, colon and liver cells, using organoid culture.
Osorio, F. G. et al. Somatic mutations reveal lineage relationships and age-related mutagenesis in human hematopoiesis. Cell Rep. 25, 2308–2316.e4 (2018).
Kuijk, E. et al. Early divergence of mutational processes in human fetal tissues. Sci. Adv. 5, eaaw1271 (2019).
Hasaart, K. A. L. et al. Mutation accumulation and developmental lineages in normal and Down syndrome human fetal haematopoiesis. Sci. Rep. 10, 12991 (2020).
Pfeifer, G. P. Mutagenesis at methylated CpG sequences. Curr. Top. Microbiol. Immunol. 301, 259–281 (2006).
Harris, R. S., Petersen-Mahrt, S. K. & Neuberger, M. S. RNA editing enzyme APOBEC1 and some of its homologs can act as DNA mutators. Mol. Cell 10, 1247–1253 (2002).
Taylor, B. J. et al. DNA deaminases induce break-associated mutation showers with implication of APOBEC3B and 3A in breast cancer kataegis. eLife 2, e00534 (2013).
Burns, M. B. et al. APOBEC3B is an enzymatic source of mutation in breast cancer. Nature 494, 366–370 (2013).
Pfeifer, G. P., You, Y. H. & Besaratinia, A. Mutations induced by ultraviolet light. Mutat. Res. 571, 19–31 (2005).
Pfeifer, G. P. et al. Tobacco smoke carcinogens, DNA damage and p53 mutations in smoking-associated cancers. Oncogene 21, 7435–7451 (2002).
Chang, J. et al. Genomic analysis of oesophageal squamous-cell carcinoma identifies alcohol drinking-related mutation signature and genomic alterations. Nat. Commun. 8, 15290 (2017).
Tomita-Mitchell, A. et al. Mismatch repair deficient human cells: spontaneous and MNNG-induced mutational spectra in the HPRT gene. Mutat. Res. 450, 125–138 (2000).
Poon, S. L. et al. Genome-wide mutational signatures of aristolochic acid and its application as a screening tool. Sci. Transl. Med. 5, 197ra101 (2013).
Letouze, E. et al. Mutational signatures reveal the dynamic interplay of risk factors and cellular processes during liver tumorigenesis. Nat. Commun. 8, 1315 (2017).
Maura, F. et al. A practical guide for mutational signature analysis in hematological malignancies. Nat. Commun. 10, 2969 (2019).
Pleguezuelos-Manzano, C. et al. Mutational signature in colorectal cancer caused by genotoxic pks+ E. coli. Nature 580, 269–273 (2020).
Lawson, A. R. J. et al. Extensive heterogeneity in somatic mutation and selection in the human bladder. Science 370, 75–82 (2020).
Li, Y. et al. Patterns of somatic structural variation in human cancer genomes. Nature 578, 112–121 (2020).
Busque, L. et al. Recurrent somatic TET2 mutations in normal elderly individuals with clonal hematopoiesis. Nat. Genet. 44, 1179–1181 (2012).
Xie, M. et al. Age-related mutations associated with clonal hematopoietic expansion and malignancies. Nat. Med. 20, 1472–1478 (2014).
McKerrell, T. et al. Leukemia-associated somatic mutations drive distinct patterns of age-related clonal hemopoiesis. Cell Rep. 10, 1239–1245 (2015).
Young, A. L., Challen, G. A., Birmann, B. M. & Druley, T. E. Clonal haematopoiesis harbouring AML-associated mutations is ubiquitous in healthy adults. Nat. Commun. 7, 12484 (2016).
Papaemmanuil, E. et al. Clinical and biological implications of driver mutations in myelodysplastic syndromes. Blood 122, 3616–3627 (2013).
Haferlach, T. et al. Landscape of genetic lesions in 944 patients with myelodysplastic syndromes. Leukemia 28, 241–247 (2014).
Papaemmanuil, E. et al. Genomic classification and prognosis in acute myeloid leukemia. N. Engl. J. Med. 374, 2209–2221 (2016).
Grinfeld, J. et al. Classification and personalized prognosis in myeloproliferative neoplasms. N. Engl. J. Med. 379, 1416–1430 (2018).
Tyner, J. W. et al. Functional genomic landscape of acute myeloid leukaemia. Nature 562, 526–531 (2018).
Laurie, C. C. et al. Detectable clonal mosaicism from birth to old age and its relationship to cancer. Nat. Genet. 44, 642–650 (2012).
Jacobs, K. B. et al. Detectable clonal mosaicism and its relationship to aging and cancer. Nat. Genet. 44, 651–658 (2012).
Loh, P. R. et al. Insights into clonal haematopoiesis from 8,342 mosaic chromosomal alterations. Nature 559, 350–355 (2018).
Suda, K. et al. Clonal expansion and diversification of cancer-associated mutations in endometriosis and normal endometrium. Cell Rep. 24, 1777–1789 (2018). This study shows that endometriosis may derive from endometrial glands that acquire driver mutations by retrograde menstruation.
Li, R. et al. Macroscopic somatic clonal expansion in morphologically normal human urothelium. Science 370, 82–89 (2020).
Olafsson, S. et al. Somatic evolution in non-neoplastic IBD-affected colon. Cell 182, 672–684.e11 (2020).
Zhu, M. et al. Somatic mutations increase hepatic clonal fitness and regeneration in chronic liver disease. Cell 177, 608–621.e12 (2019).
Inman, G. J. et al. The genomic landscape of cutaneous SCC reveals drivers and a novel azathioprine associated mutational signature. Nat. Commun. 9, 3667 (2018).
Bonilla, X. et al. Genomic analysis identifies new drivers and progression pathways in skin basal cell carcinoma. Nat. Genet. 48, 398–406 (2016).
Besaratinia, A., Kim, S. I., Bates, S. E. & Pfeifer, G. P. Riboflavin activated by ultraviolet A1 irradiation induces oxidative DNA damage-mediated mutations inhibited by vitamin C. Proc. Natl Acad. Sci. USA 104, 5953–5958 (2007).
Yizhak, K. et al. RNA sequence analysis reveals macroscopic somatic clonal expansion across normal tissues. Science 364, eaaw0726 (2019).
Totoki, Y. et al. Trans-ancestry mutational landscape of hepatocellular carcinoma genomes. Nat. Genet. 46, 1267–1273 (2014).
Alberg, A. J., Brock, M. V., Ford, J. G., Samet, J. M. & Spivack, S. D. Epidemiology of lung cancer: diagnosis and management of lung cancer, 3rd ed: American College of Chest Physicians evidence-based clinical practice guidelines. Chest 143, e1S–e29S (2013).
Tang, J. et al. The genomic landscapes of individual melanocytes from human skin. Nature 586, 600–605 (2020).
Kang, H. & Shibata, D. Direct measurements of human colon crypt stem cell niche genetic fidelity: the role of chance in non-Darwinian mutation selection. Front. Oncol. 3, 264 (2013).
Nguyen, H. et al. Deficient Pms2, ERCC1, Ku86, CcOI in field defects during progression to colon cancer. J. Vis. Exp. 41, e1931 (2010).
Buc, E. et al. High prevalence of mucosa-associated E. coli producing cyclomodulin and genotoxin in colon cancer. PLoS ONE 8, e56964 (2013).
Salk, J. J. et al. Clonal expansions in ulcerative colitis identify patients with neoplasia. Proc. Natl Acad. Sci. USA 106, 20871–20876 (2009).
Stachler, M. D. et al. Detection of mutations in Barrett’s esophagus before progression to high-grade dysplasia or adenocarcinoma. Gastroenterology 155, 156–167 (2018).
Fujimoto, A. et al. Whole-genome sequencing of liver cancers identifies etiological influences on mutation patterns and recurrent mutations in chromatin regulators. Nat. Genet. 44, 760–764 (2012).
Schulze, K. et al. Exome sequencing of hepatocellular carcinomas identifies new mutational signatures and potential therapeutic targets. Nat. Genet. 47, 505–511 (2015).
Cancer Genome Atlas Research Network. Comprehensive and integrative genomic characterization of hepatocellular carcinoma. Cell 169, 1327–1341.e23 (2017).
Kensler, T. W. et al. Transforming cancer prevention through precision medicine and immune-oncology. Cancer Prev. Res. 9, 2–10 (2016).
Curtius, K., Wright, N. A. & Graham, T. A. An evolutionary perspective on field cancerization. Nat. Rev. Cancer 18, 19–32 (2018).
Hori, M. et al. Cancer incidence and incidence rates in Japan in 2009: a study of 32 population-based cancer registries for the Monitoring of Cancer Incidence in Japan (MCIJ) project. Jpn. J. Clin. Oncol. 45, 884–891 (2015).
Statistical Research and Applications Branch, National Cancer Institute. DevCan: Probability of Developing or Dying of Cancer Software. Version 6.7.7 (National Cancer Institute, 2019).
Abelson, S. et al. Prediction of acute myeloid leukaemia risk in healthy individuals. Nature 559, 400–404 (2018).
Desai, P. et al. Somatic mutations precede acute myeloid leukemia years before diagnosis. Nat. Med. 24, 1015–1023 (2018).
Akbari, M. R. et al. PPM1D mutations in circulating white blood cells and the risk for ovarian cancer. J. Natl Cancer Inst. 106, djt323 (2014).
Wong, T. N. et al. Cellular stressors contribute to the expansion of hematopoietic clones of varying leukemic potential. Nat. Commun. 9, 455 (2018).
Sun, X. et al. Arid1a has context-dependent oncogenic and tumor suppressor functions in liver cancer. Cancer Cell 32, 574–589.e6 (2017).
Alcolea, M. P. et al. Differentiation imbalance in single oesophageal progenitor cells causes clonal immortalization and field change. Nat. Cell Biol. 16, 615–622 (2014).
Demehri, S., Turkoz, A. & Kopan, R. Epidermal Notch1 loss promotes skin tumorigenesis by impacting the stromal microenvironment. Cancer Cell 16, 55–66 (2009).
Kagoya, Y. et al. JAK2V617F+ myeloproliferative neoplasm clones evoke paracrine DNA damage to adjacent normal cells through secretion of lipocalin-2. Blood 124, 2996–3006 (2014).
Jaiswal, S. et al. Clonal hematopoiesis and risk of atherosclerotic cardiovascular disease. N. Engl. J. Med. 377, 111–121 (2017).
Fuster, J. J. et al. Clonal hematopoiesis associated with TET2 deficiency accelerates atherosclerosis development in mice. Science 355, 842–847 (2017).
Qian, Y. et al. The adaptor Act1 is required for interleukin 17-dependent signaling associated with autoimmune and inflammatory disease. Nat. Immunol. 8, 247–256 (2007).
Lee, J. S. et al. Interleukin-23-independent IL-17 production regulates intestinal epithelial permeability. Immunity 43, 727–738 (2015).
Reikvam, D. H. et al. Epithelial–microbial crosstalk in polymeric Ig receptor deficient mice. Eur. J. Immunol. 42, 2959–2970 (2012).
Joseph, C. G. et al. Association of the autoimmune disease scleroderma with an immunologic response to cancer. Science 343, 152–157 (2014).
Olcaydu, D. et al. A common JAK2 haplotype confers susceptibility to myeloproliferative neoplasms. Nat. Genet. 41, 450–454 (2009).
Kilpivaara, O. et al. A germline JAK2 SNP is associated with predisposition to the development of JAK2(V617F)-positive myeloproliferative neoplasms. Nat. Genet. 41, 455–459 (2009).
Jones, A. V. et al. JAK2 haplotype is a major risk factor for the development of myeloproliferative neoplasms. Nat. Genet. 41, 446–449 (2009).
Flavahan, W. A., Gaskell, E. & Bernstein, B. E. Epigenetic plasticity and the hallmarks of cancer. Science 357, eaal2380 (2017).
Hanahan, D. & Weinberg, R. A. Hallmarks of cancer: the next generation. Cell 144, 646–674 (2011).
Wu, J. & Izpisua Belmonte, J. C. Stem cells: a renaissance in human biology research. Cell 165, 1572–1585 (2016).
Atlasi, Y. & Stunnenberg, H. G. The interplay of epigenetic marks during stem cell differentiation and development. Nat. Rev. Genet. 18, 643–658 (2017).
Coorens, T. H. H. et al. Embryonal precursors of Wilms tumor. Science 366, 1247–1251 (2019). This study describes that H19 locus hypermethylation, epi-mutation, can drive clonal expansion during nephrogenesis, which can contribute to the development of Wilms tumour.
Khurana, E. et al. Integrative annotation of variants from 1092 humans: application to cancer genomics. Science 342, 1235587 (2013).
Fredriksson, N. J., Ny, L., Nilsson, J. A. & Larsson, E. Systematic analysis of noncoding somatic mutations and gene expression alterations across 14 tumor types. Nat. Genet. 46, 1258–1263 (2014).
Northcott, P. A. et al. Enhancer hijacking activates GFI1 family oncogenes in medulloblastoma. Nature 511, 428–434 (2014).
Weinhold, N., Jacobsen, A., Schultz, N., Sander, C. & Lee, W. Genome-wide analysis of noncoding regulatory mutations in cancer. Nat. Genet. 46, 1160–1165 (2014).
Melton, C., Reuter, J. A., Spacek, D. V. & Snyder, M. Recurrent somatic mutations in regulatory regions of human cancer genomes. Nat. Genet. 47, 710–716 (2015).
Puente, X. S. et al. Non-coding recurrent mutations in chronic lymphocytic leukaemia. Nature 526, 519–524 (2015).
Nik-Zainal, S. et al. Landscape of somatic mutations in 560 breast cancer whole-genome sequences. Nature 534, 47–54 (2016).
Imielinski, M., Guo, G. & Meyerson, M. Insertions and deletions target lineage-defining genes in human cancers. Cell 168, 460–472.e14 (2017).
Rheinbay, E. et al. Analyses of non-coding somatic drivers in 2,658 cancer whole genomes. Nature 578, 102–111 (2020).
Pickar-Oliver, A. & Gersbach, C. A. The next generation of CRISPR–Cas technologies and applications. Nat. Rev. Mol. Cell Biol. 20, 490–507 (2019).
Kretzschmar, K. & Clevers, H. Organoids: modeling development and the stem cell niche in a dish. Dev. Cell 38, 590–600 (2016).
Drost, J. & Clevers, H. Organoids in cancer research. Nat. Rev. Cancer 18, 407–418 (2018).
Burgess, D. J. Spatial transcriptomics coming of age. Nat. Rev. Genet. 20, 317 (2019).
Stuart, T. & Satija, R. Integrative single-cell analysis. Nat. Rev. Genet. 20, 257–272 (2019).
Schatz, M. C. Nanopore sequencing meets epigenetics. Nat. Methods 14, 347–348 (2017).
Ho, S. S., Urban, A. E. & Mills, R. E. Structural variation in the sequencing era. Nat. Rev. Genet. 21, 171–189 (2020).
Jaiswal, S. & Ebert, B. L. Clonal hematopoiesis in human aging and disease. Science 366, eaan4673 (2019).
Freed, D., Stevens, E. L. & Pevsner, J. Somatic mosaicism in the human genome. Genes 5, 1064–1094 (2014).
Linder, D. & Gartler, S. M. Distribution of glucose-6-phosphate dehydrogenase electrophoretic variants in different tissues of heterozygotes. Am. J. Hum. Genet. 17, 212–220 (1965).
Behjati, S. et al. Genome sequencing of normal cells reveals developmental lineages and mutational processes. Nature 513, 422–425 (2014).
Ju, Y. S. et al. Somatic mutations reveal asymmetric cellular dynamics in the early human embryo. Nature 543, 714–718 (2017).
Hou, Y. et al. Single-cell exome sequencing and monoclonal evolution of a JAK2-negative myeloproliferative neoplasm. Cell 148, 873–885 (2012).
Nicholson, A. M. et al. Fixation and spread of somatic mutations in adult human colonic epithelium. Cell Stem Cell 22, 909–918.e8 (2018).
Author information
Authors and Affiliations
Contributions
The authors contributed equally to all aspects of the article.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information
Nature Reviews Cancer thanks R. van Boxtel and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Glossary
- Driver mutations
-
Those mutations that confer selective advantage for their target cells to allow clonal outgrowth, whereas those mutations acquired prior to a driver mutation are also inherited by driver-mutated and clonally expanded cells despite conferring no selective roles and are called passenger mutations.
- Positively selected clones
-
Clones that outcompete surrounding cells by acquisition of driver mutations that confer selective advantages to the cells.
- Lyonized cells
-
Those cells undergoing inactivation of one of the two X chromosome copies in females.
- Skewed chromosome X inactivation
-
In the presence of a clone, inactivation of one of the two chromosomes in early development of female organisms (lyonization) may be skewed from the 1:1 ratio that is expected for a random process, which can be used to infer the presence of clonal evolution in the sample of interest.
- Genetic drift
-
A change in the frequency of an existing gene variant in a population due to random sampling of cells or organisms.
- Allele dropout
-
The preferential amplification of one of a pair of heterozygous alleles frequently observed in amplification from a single cell or a small number of cells, in which the other allele is totally under-represented or fails to be detected.
- Ratio of non-synonymous to synonymous mutations
-
(dN/dS). The ratio of the number of non-synonymous substitutions per non-synonymous site to the number of synonymous substitutions per synonymous site, frequently used as an indicator of selective pressure acting on a protein coding gene.
- Variant allele frequency
-
The frequency of the mutated allele, correlating with the clonal cell fraction within a sample, that is calculated by dividing the number of mutant reads by the total number of reads that encompass the mutated position in sequencing analysis.
- Loss of heterozygosity
-
An allelic state in which one of the two parental alleles is lost with or without copy number loss, providing a common mechanism of biallelic inactivation of tumour suppressor genes or duplication of a gain-of-function mutation accompanied by loss of wild-type alleles.
- Parity
-
The number of times a female has given birth.
- Chromothripsis
-
A mutational process by which clustered chromosomal rearrangements occur in a single event affecting one or a few chromosomes.
- Negative selection
-
The selective removal of defective/disadvantageous alleles or cells having such alleles from a population of species or cells.
- Field carcinogenesis
-
A biological process in which large areas of tissues or organs are affected by carcinogenic alterations.
- Epi-mutations
-
Heritable changes without any accompanying actual base pair changes in primary DNA sequences, which are typically represented by DNA methylation, histone modifications and other mechanisms.
Rights and permissions
About this article
Cite this article
Kakiuchi, N., Ogawa, S. Clonal expansion in non-cancer tissues. Nat Rev Cancer 21, 239–256 (2021). https://doi.org/10.1038/s41568-021-00335-3
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41568-021-00335-3
This article is cited by
-
Aneuploid embryonic stem cells drive teratoma metastasis
Nature Communications (2024)
-
A novel bioinformatic approach reveals cooperation between Cancer/Testis genes in basal-like breast tumors
Oncogene (2024)
-
Tissue mosaicism following stem cell aging: blood as an exemplar
Nature Aging (2024)
-
A timeline of tumour-associated macrophage biology
Nature Reviews Cancer (2023)
-
An ATR-PrimPol pathway confers tolerance to oncogenic KRAS-induced and heterochromatin-associated replication stress
Nature Communications (2023)