Abstract
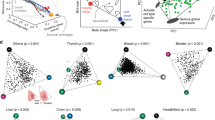
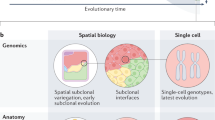
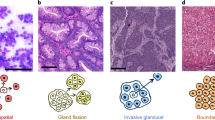
Tumours vary in gene expression programmes and genetic alterations. Understanding this diversity and its biological meaning requires a theoretical framework, which could in turn guide the development of more accurate prognosis and therapy. Here, we review the theory of multi-task evolution of cancer, which is based upon the premise that tumours evolve in the host and face selection trade-offs between multiple biological functions. This theory can help identify the major biological tasks that cancer cells perform and the trade-offs between these tasks. It introduces the concept of specialist tumours, which focus on one task, and generalist tumours, which perform several tasks. Specialist tumours are suggested to be sensitive to therapy targeting their main task. Driver mutations tune gene expression towards specific tasks in a tissue-dependent manner and thus help to determine whether a tumour is specialist or generalist. We discuss potential applications of the theory of multi-task evolution to interpret the spatial organization of tumours and intratumour heterogeneity.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Cancer Genome Atlas Research Network et al. Integrated genomic characterization of papillary thyroid carcinoma. Cell 159, 676–690 (2014).
Cancer Genome Atlas Network. Comprehensive molecular portraits of human breast tumours. Nature 490, 61–70 (2012).
Curtis, C. et al. The genomic and transcriptomic architecture of 2,000 breast tumours reveals novel subgroups. Nature 486, 346–352 (2012).
Eirew, P. et al. Dynamics of genomic clones in breast cancer patient xenografts at single-cell resolution. Nature 518, 422–426 (2015).
Keren, L. et al. A structured tumor-immune microenvironment in triple negative breast cancer revealed by multiplexed ion beam imaging. Cell 174, 1373–1387.e19 (2018).
Kim, C. et al. Chemoresistance evolution in triple-negative breast cancer delineated by single-cell sequencing. Cell 173, 879–893.e13 (2018).
Li, H. et al. Reference component analysis of single-cell transcriptomes elucidates cellular heterogeneity in human colorectal tumors. Nat. Genet. 49, 708–718 (2017).
Ling, S. et al. Extremely high genetic diversity in a single tumor points to prevalence of non-Darwinian cell evolution. Proc. Natl Acad. Sci. USA 112, E6496–E6505 (2015).
McGranahan, N. & Swanton, C. Clonal heterogeneity and tumor evolution: past, present, and the future. Cell 168, 613–628 (2017).
Navin, N. et al. Tumour evolution inferred by single-cell sequencing. Nature 472, 90–94 (2011).
Tirosh, I. et al. Dissecting the multicellular ecosystem of metastatic melanoma by single-cell RNA-seq. Science 352, 189–196 (2016).
Venteicher, A. S. et al. Decoupling genetics, lineages, and microenvironment in IDH-mutant gliomas by single-cell RNA-seq. Science 355, eaai8478 (2017).
Hu, Z., Sun, R. & Curtis, C. A population genetics perspective on the determinants of intra-tumor heterogeneity. Biochim. Biophys. Acta Rev. Cancer 1867, 109–126 (2017).
Lipinski, K. A. et al. Cancer evolution and the limits of predictability in precision cancer medicine. Trends Cancer 2, 49–63 (2016).
Vogelstein, B. et al. Cancer genome landscapes. Science 339, 1546–1558 (2013).
Nagpal, K. et al. Development and validation of a deep learning algorithm for improving Gleason scoring of prostate cancer. NPJ Digit. Med. 2, 48 (2019).
Maley, C. C. et al. Genetic clonal diversity predicts progression to esophageal adenocarcinoma. Nat. Genet. 38, 468–473 (2006).
Maley, C. C., Koelble, K., Natrajan, R., Aktipis, A. & Yuan, Y. An ecological measure of immune-cancer colocalization as a prognostic factor for breast cancer. Breast Cancer Res. 17, 131 (2015).
Mobadersany, P. et al. Predicting cancer outcomes from histology and genomics using convolutional networks. Proc. Natl Acad. Sci. USA 115, E2970–E2979 (2018).
Robertson, S., Azizpour, H., Smith, K. & Hartman, J. Digital image analysis in breast pathology — from image processing techniques to artificial intelligence. Transl. Res. 194, 19–35 (2018).
Sørlie, T. et al. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc. Natl Acad. Sci. USA 98, 10869–10874 (2001).
Beerenwinkel, N., Greenman, C. D. & Lageren, J. Computational cancer biology: an evolutionary perspective. PLoS Comput. Biol. 12, e1004717 (2016).
Gatenby, R. A. & Brown, J. Mutations, evolution and the central role of a self-defined fitness function in the initiation and progression of cancer. Biochim. Biophys. Acta Rev. Cancer 1867, 162–166 (2017).
Greaves, M. & Maley, C. C. Clonal evolution in cancer. Nature 481, 306–313 (2012).
Nowell, P. C. The clonal evolution of tumor cell populations. Science 194, 23–28 (1976).
Sottoriva, A., Barnes, C. P. & Graham, T. A. Catch my drift? Making sense of genomic intra-tumour heterogeneity. Biochim. Biophys. Acta Rev. Cancer 1867, 95–100 (2017).
Durinck, S. et al. Temporal dissection of tumorigenesis in primary cancers. Cancer Discov. 1, 137–143 (2011).
Schwartz, R. & Schäffer, A. A. The evolution of tumour phylogenetics: principles and practice. Nat. Rev. Genet. 18, 213–229 (2017).
Martincorena, I. et al. Universal patterns of selection in cancer and somatic tissues. Cell 171, 1029–1041.e21 (2017).
Williams, M. J., Werner, B., Barnes, C. P., Graham, T. A. & Sottoriva, A. Identification of neutral tumor evolution across cancer types. Nat. Genet. 48, 238–244 (2016).
Gillespie, J. H. Population Genetics: A Concise Guide (Johns Hopkins University Press, 2004).
McDonald, T. O., Chakrabarti, S. & Michor, F. Currently available bulk sequencing data do not necessarily support a model of neutral tumor evolution. Nat. Genet. 50, 1620–1623 (2018).
Sottoriva, A. et al. A big bang model of human colorectal tumor growth. Nat. Genet. 47, 209–216 (2015).
Tarabichi, M. et al. Neutral tumor evolution? Nat. Genet. 50, 1630–1633 (2018).
Waclaw, B. et al. A spatial model predicts that dispersal and cell turnover limit intratumour heterogeneity. Nature 525, 261–264 (2015).
Alon, U. An Introduction to Systems Biology: Design Principles of Biological Circuits (CRC Press, 2019).
Shoval, O. et al. Evolutionary trade-offs, pareto optimality, and the geometry of phenotype space. Science 336, 1157–1160 (2012).
Hanahan, D. & Weinberg, R. A. Hallmarks of cancer: the next generation. Cell 144, 646–674 (2011).
Aktipis, C. A., Boddy, A. M., Gatenby, R. A., Brown, J. S. & Maley, C. C. Life history trade-offs in cancer evolution. Nat. Rev. Cancer 13, 883–892 (2013).
Gallaher, J. A., Brown, J. S. & Anderson, A. R. A. The impact of proliferation-migration tradeoffs on phenotypic evolution in cancer. Sci. Rep. 9, 1–10 (2019).
Gillies, R. J., Brown, J. S., Anderson, A. R. A. & Gatenby, R. A. Eco-evolutionary causes and consequences of temporal changes in intratumoural blood flow. Nat. Rev. Cancer 18, 576–585 (2018).
Hatzikirou, H., Basanta, D., Simon, M., Schaller, K. & Deutsch, A. ‘Go or grow’: the key to the emergence of invasion in tumour progression? Math. Med. Biol. 29, 49–65 (2012).
Broxterman, H. J. et al. Induction by verapamil of a rapid increase in ATP consumption in multidrug-resistant tumor cells. FASEB J. 2, 2278–2282 (1988).
Jerby, L. et al. Metabolic associations of reduced proliferation and oxidative stress in advanced breast cancer. Cancer Res. 72, 5712–5720 (2012).
Gade, T. P. F. et al. Ischemia induces quiescence and autophagy dependence in hepatocellular carcinoma. Radiology 283, 702–710 (2017).
Wang, X. et al. Exit from quiescence displays a memory of cell growth and division. Nat. Commun. 8, 321 (2017).
Evdokimova, V., Tognon, C., Ng, T. & Sorensen, P. H. Reduced proliferation and enhanced migration: two sides of the same coin? Molecular mechanisms of metastatic progression by YB-1. Cell Cycle 8, 2901–2906 (2009).
Tsai, J. H. & Yang, J. Epithelial-mesenchymal plasticity in carcinoma metastasis. Genes Dev. 27, 2192–2206 (2013).
Orlando, P. A., Gatenby, R. A. & Brown, J. S. Tumor evolution in space: the effects of competition colonization tradeoffs on tumor invasion dynamics. Front. Oncol. 3, 45 (2013).
Adler, M. et al. Continuum of gene-expression profiles provides spatial division of labor within a differentiated cell type article continuum of gene-expression profiles provides spatial division of labor within a differentiated cell type. Cell Syst. 8, 43–52.e5 (2019).
Hart, Y. et al. Inferring biological tasks using Pareto analysis of high-dimensional data. Nat. Methods 12, 233–235 (2015).
Korem, Y. et al. Geometry of the gene expression space of individual cells. PLoS Comput. Biol. 11, e1004224 (2015).
Szekely, P., Sheftel, H., Mayo, A. & Alon, U. Evolutionary tradeoffs between economy and effectiveness in biological homeostasis systems. PLoS Comput. Biol. 9, e1003163 (2013).
Tendler, A., Mayo, A. & Alon, U. Evolutionary tradeoffs, Pareto optimality and the morphology of ammonite shells. BMC Syst. Biol. 9, 12 (2015).
Szekely, P., Korem, Y., Moran, U., Mayo, A. & Alon, U. The mass-longevity triangle: pareto optimality and the geometry of life-history trait space. PLoS Comput. Biol. 11, 1–19 (2015).
Hausser, J. et al. Tumor diversity and the trade-off between universal cancer tasks. Nat. Commun. 10, 5423 (2019).
Trink, A. et al. Geometry of gene expression space of Wilms’ tumors from human patients. Neoplasia 20, 871–881 (2018).
Cutler, A. A. & Breiman, L. Archetypial analysis. Technometrics 36, 338–347 (1994).
Mørup, M. & Hansen, L. K. Archetypal analysis for machine learning and data mining. Neurocomputing 80, 54–63 (2012).
Aubert, O. et al. Archetype analysis identifies distinct profiles in renal transplant recipients with transplant glomerulopathy associated with allograft survival. J. Am. Soc. Nephrol. 30, 625–639 (2019).
Dai, Z. et al. Identification of cancer-associated metabolic vulnerabilities by modeling multi-objective optimality in metabolism. Cell Commun. Signal. 17, 124 (2019).
Thøgersen, J. C., Mørup, M., Damkiær, S., Molin, S. & Jelsbak, L. Archetypal analysis of diverse Pseudomonas aeruginosa transcriptomes reveals adaptation in cystic fibrosis airways. BMC Bioinformatics 14, 279 (2013).
van Dijk, D. et al. Recovering gene interactions from single-cell data using data diffusion. Cell 174, 716–729.e27 (2018).
Wei, S. C. et al. Negative co-stimulation constrains T cell differentiation by imposing boundaries on possible cell states. Immunity 50, 1084–1098.e10 (2019).
Arnold, S. J. Morphology, performance and fitness. Am. Zool. 23, 347–361 (1983).
Basan, M. et al. Overflow metabolism in Escherichia coli results from efficient proteome allocation. Nature 528, 99–104 (2015).
Scott, M., Gunderson, C. W., Mateescu, E. M., Zhang, Z. & Hwa, T. Interdependence of cell growth and gene expression: origins and consequences. Science 330, 1099–1102 (2010).
Towbin, B. D. et al. Optimality and sub-optimality in a bacterial growth law. Nat. Commun. 8, 14123 (2017).
Reding-Roman, C. et al. The unconstrained evolution of fast and efficient antibiotic-resistant bacterial genomes. Nat. Ecol. Evol. https://doi.org/10.1038/s41559-016-0050 (2017).
Vander Heiden, M. G., Cantley, L. C. & Thompson, C. B. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science 324, 1029–1033 (2009).
Rueffler, C., Hermisson, J. & Wagner, G. P. Evolution of functional specialization and division of labor. Proc. Natl Acad. Sci. USA 109, E326–E335 (2012).
Sheftel, H., Shoval, O., Mayo, A. & Alon, U. The geometry of the Pareto front in biological phenotype space. Ecol. Evol. 3, 1471–1483 (2013).
Gillies, R. J., Verduzco, D. & Gatenby, R. A. Evolutionary dynamics of carcinogenesis and why targeted therapy does not work. Nat. Rev. Cancer 12, 487–493 (2012).
Halpern, K. B. et al. Single-cell spatial reconstruction reveals global division of labour in the mammalian liver. Nature 542, 352–356 (2017).
Gatenby, R. A. & Gillies, R. J. Why do cancers have high aerobic glycolysis? Nat. Rev. Cancer 4, 891–899 (2004).
Sonveaux, P. et al. Targeting lactate-fueled respiration selectively kills hypoxic tumor cells in mice. J. Clin. Invest. 118, 3930–3942 (2008).
Kumar, S. et al. Intra-tumoral metabolic zonation and resultant phenotypic diversification are dictated by blood vessel proximity. Cell Metab. 30, 201–211.e6 (2019).
Dewhirst, M. W. & Secomb, T. W. Transport of drugs from blood vessels to tumour tissue. Nat. Rev. Cancer 17, 738–750 (2017).
McGranahan, N. et al. Clonal status of actionable driver events and the timing of mutational processes in cancer evolution. Sci. Transl. Med. 7, 283ra54 (2015).
Bioucas-Dias, J. A Variable Splitting Augmented Lagrangian Approach to Linear Spectral Unmixing. (IEEE, 2009). https://doi.org/10.1109/WHISPERS.2009.5289072
Kanehisa, M., Sato, Y., Furumichi, M., Morishima, K. & Tanabe, M. New approach for understanding genome variations in KEGG. Nucleic Acids Res. 47, D590–D595 (2019).
Fabregat, A. et al. The reactome pathway knowledgebase. Nucleic Acids Res. 46, D649–D655 (2018).
Subramanian, A. et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl Acad. Sci. USA 102, 15545–15550 (2005).
van Dijk, D. et al. Finding Archetypal Spaces for Data Using Neural Networks (Cornell University, 2019).
Laplante, M. & Sabatini, D. M. mTOR signaling in growth control and disease. Cell 149, 274–293 (2012).
Azizi, E. et al. Single-cell map of diverse immune phenotypes in the breast tumor microenvironment. Cell 174, 1293–1308 (2018).
Crosetto, N., Bienko, M. & van Oudenaarden, A. Spatially resolved transcriptomics and beyond. Nat. Rev. Genet. 16, 57–66 (2015).
Lein, E., Borm, L. E. & Linnarsson, S. The promise of spatial transcriptomics for neuroscience in the era of molecular cell typing. Science 358, 64–69 (2017).
Moor, A. E. et al. Global mRNA polarization regulates translation efficiency in the intestinal epithelium. Science 357, 1299–1303 (2017).
Satija, R., Farrell, J. A., Gennert, D., Schier, A. F. & Regev, A. Spatial reconstruction of single-cell gene expression data. Nat. Biotechnol. 33, 495–502 (2015).
Stahl, P. L. et al. Visualization and analysis of gene expression in tissue sections by spatial transcriptomics. Science 353, 78–82 (2016).
Wang, H. A. et al. Fast chemical imaging at high spatial resolution by laser ablation inductively coupled plasma mass spectrometry. Anal. Chem. 85, 10107–10116 (2013).
Ke, R. et al. In situ sequencing for RNA analysis in preserved tissue and cells. Nat. Methods 10, 857–860 (2013).
Chen, K. H., Boettiger, A. N., Moffitt, J. R., Wang, S. & Zhuang, X. Spatially resolved, highly multiplexed RNA profiling in single cells. Science 348, aaa6090 (2015).
Eng, C. L. et al. Transcriptome-scale super-resolved imaging in tissues by RNA seqFISH. Nature 568, 235–239 (2019).
Nagrath, D. et al. Integrated energy and flux balance based multiobjective framework for large-scale metabolic networks. Ann. Biomed. Eng. 35, 863–885 (2007).
Gebhardt, R. & Matz-Soja, M. Liver zonation: novel aspects of its regulation and its impact on homeostasis. World J. Gastroenterol. 20, 8491 (2014).
Junttila, M. R. & De Sauvage, F. J. Influence of tumour micro-environment heterogeneity on therapeutic response. Nature 501, 346–354 (2013).
Maley, C. C. et al. Classifying the evolutionary and ecological features of neoplasms. Nat. Rev. Cancer 17, 605–619 (2017).
Nawaz, S., Heindl, A., Koelble, K. & Yuan, Y. Beyond immune density: critical role of spatial heterogeneity in estrogen receptor-negative breast cancer. Mod. Pathol. 28, 766–777 (2015).
Yuan, Y. et al. Quantitative Image Analysis of Cellular Heterogeneity in Breast Tumors Complements Genomic Profiling. Sci. Transl. Med. 4, 157ra143 (2012).
Lloyd, M. C. et al. Darwinian dynamics of intratumoral heterogeneity: not solely random mutations but also variable environmental selection forces. Cancer Res. 76, 3136–3144 (2016).
Lloyd, M. C. et al. Vascular measurements correlate with estrogen receptor status. BMC Cancer 14, 279 (2014).
Carmona-Fontaine, C. et al. Emergence of spatial structure in the tumor microenvironment due to the Warburg effect. Proc. Natl Acad. Sci. USA 110, 19402–19407 (2013).
Carmona-Fontaine, C. et al. Metabolic origins of spatial organization in the tumor microenvironment. Proc. Natl Acad. Sci. USA 114, 2934–2939 (2017).
Tredan, O., Galmarini, C. M., Patel, K. & Tannock, I. F. Drug resistance and the solid tumor microenvironment. J. Natl Cancer Inst. 99, 1441–1454 (2007).
Anderson, A. R. A., Weaver, A. M., Cummings, P. T. & Quaranta, V. Tumor morphology and phenotypic evolution driven by selective pressure from the microenvironment. Cell 127, 905–915 (2006).
Li, X. et al. Temporal and spatial evolution of somatic chromosomal alterations: a case-cohort study of barrett’s esophagus. Cancer Prev. Res. 7, 114–127 (2014).
Lin, J. R. et al. Highly multiplexed immunofluorescence imaging of human tissues and tumors using t-CyCIF and conventional optical microscopes. eLife 7, e31657 (2018).
Rosenbloom, D. I. S., Camara, P. G., Chu, T. & Rabadan, R. Evolutionary scalpels for dissecting tumor ecosystems. Biochim. Biophys. Acta Rev. Cancer 1867, 69–83 (2017).
Azmi, A. S., Bao, B. & Sarkar, F. H. Exosomes in cancer development, metastasis, and drug resistance: a comprehensive review. Cancer Metastasis Rev. 32, 623–642 (2013).
Jones, V. S. et al. Cytokines in cancer drug resistance: cues to new therapeutic strategies. Biochim. Biophys. Acta Rev. Cancer 1865, 255–265 (2016).
Tabassum, D. P. & Polyak, K. Tumorigenesis: it takes a village. Nat. Rev. Cancer 15, 473–483 (2015).
Chapman, A. et al. Heterogeneous tumor subpopulations cooperate to drive invasion. Cell Rep. 8, 688–695 (2014).
Acknowledgements
The authors thank R. Shouval and members of U.A. and J. Joyce’s labs for feedback on the manuscript. This work was supported by the Minerva foundation. U.A. is the incumbent of the Abisch-Frenkel Professorial Chair and acknowledges funding by BSF-NSF-NIH-CRCNS. J.H. acknowledges the support of the Swiss National Science Foundation (#177868), the Swedish Research Council and the Swedish Cancer Society.
Author information
Authors and Affiliations
Contributions
The authors contributed equally to all aspects of the article.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information
Nature Reviews Cancer thanks A. Anderson, T. Graham and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Related links
Pareto Task Inference software package, Matlab implementation: https://www.weizmann.ac.il/mcb/UriAlon/download/ParTI
An implementation of Pareto Task Inference for the R software: https://github.com/vitkl/ParetoTI
The IMAXT Grand Challenge: https://www.cruk.cam.ac.uk/research-groups/imaxt-laboratory/the-project
Rights and permissions
About this article
Cite this article
Hausser, J., Alon, U. Tumour heterogeneity and the evolutionary trade-offs of cancer. Nat Rev Cancer 20, 247–257 (2020). https://doi.org/10.1038/s41568-020-0241-6
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41568-020-0241-6
This article is cited by
-
Regulation and signaling pathways in cancer stem cells: implications for targeted therapy for cancer
Molecular Cancer (2023)
-
Breast cancer heterogeneity and its implication in personalized precision therapy
Experimental Hematology & Oncology (2023)
-
Lifting the curtain on molecular differences between malignant pleural mesotheliomas
Nature Genetics (2023)
-
Clonal evolution in leukemia: preleukemia, evolutionary models, and clinical implications
Genome Instability & Disease (2023)
-
Standard aberration: cancer biology and the modeling account of normal function
Biology & Philosophy (2023)