Abstract
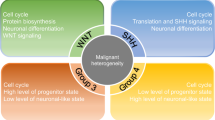
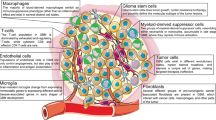
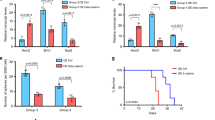
Medulloblastoma, a malignant brain tumour primarily diagnosed during childhood, has recently been the focus of intensive molecular profiling efforts, profoundly advancing our understanding of biologically and clinically heterogeneous disease subgroups. Genomic, epigenomic, transcriptomic and proteomic landscapes have now been mapped for an unprecedented number of bulk samples from patients with medulloblastoma and, more recently, for single medulloblastoma cells. These efforts have provided pivotal new insights into the diverse molecular mechanisms presumed to drive tumour initiation, maintenance and recurrence across individual subgroups and subtypes. Translational opportunities stemming from this knowledge are continuing to evolve, providing a framework for improved diagnostic and therapeutic interventions. In this Review, we summarize recent advances derived from this continued molecular characterization of medulloblastoma and contextualize this progress towards the deployment of more effective, molecularly informed treatments for affected patients.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Ostrom, Q. T. et al. CBTRUS Statistical Report: primary brain and other central nervous system tumors diagnosed in the United States in 2011–2015. Neuro-Oncology 20, iv1–iv86 (2018).
Curtin, S. C., Minino, A. M. & Anderson, R. N. Declines in cancer death rates among children and adolescents in the United States, 1999-2014. NCHS Data Brief, 1–8 (2016).
Ezzat, S. et al. Pediatric brain tumors in a low/middle income country: does it differ from that in developed world? J. Neurooncol. 126, 371–376 (2016).
Makino, K., Nakamura, H., Yano, S., Kuratsu, J. & Kumamoto Brain Tumor Group. Population-based epidemiological study of primary intracranial tumors in childhood. Childs Nerv. Syst. 26, 1029–1034 (2010).
Taylor, M. D. et al. Molecular subgroups of medulloblastoma: the current consensus. Acta Neuropathol. 123, 465–472 (2012).
Northcott, P. A. et al. Medulloblastomics: the end of the beginning. Nat. Rev. Cancer 12, 818–834 (2012).
Rausch, T. et al. Genome sequencing of pediatric medulloblastoma links catastrophic DNA rearrangements with TP53 mutations. Cell 148, 59–71 (2012). This first whole-genome sequencing study of MB identifies the association between TP53 mutations and chromothripsis in SHH MB.
Pugh, T. J. et al. Medulloblastoma exome sequencing uncovers subtype-specific somatic mutations. Nature 488, 106–110 (2012).
Northcott, P. A. et al. Subgroup-specific structural variation across 1,000 medulloblastoma genomes. Nature 488, 49–56 (2012).
Jones, D. T. et al. Dissecting the genomic complexity underlying medulloblastoma. Nature 488, 100–105 (2012).
Northcott, P. A. et al. Medulloblastoma. Nat. Rev. Dis. Primers 5, 11 (2019).
Pomeroy, S. L. et al. Prediction of central nervous system embryonal tumour outcome based on gene expression. Nature 415, 436–442 (2002).
Lee, Y. et al. A molecular fingerprint for medulloblastoma. Cancer Res. 63, 5428–5437 (2003).
Cho, Y. J. et al. Integrative genomic analysis of medulloblastoma identifies a molecular subgroup that drives poor clinical outcome. J. Clin. Oncol. 29, 1424–1430 (2011).
Kool, M. et al. Integrated genomics identifies five medulloblastoma subtypes with distinct genetic profiles, pathway signatures and clinicopathological features. PLOS ONE 3, e3088 (2008).
Northcott, P. A. et al. Medulloblastoma comprises four distinct molecular variants. J. Clin. Oncol. 29, 1408–1414 (2011).
Thompson, M. C. et al. Genomics identifies medulloblastoma subgroups that are enriched for specific genetic alterations. J. Clin. Oncol. 24, 1924–1931 (2006). Cho et al. (2011), Kool et al. (2008), Northcott et al. (2011) and Thompson et al. (2006) report the first separation of MB into distinct molecular subgroups using array-based gene expression profiling and unsupervised clustering.
Louis, D. N. et al. The 2016 World Health Organization classification of tumors of the central nervous system: a summary. Acta Neuropathol. 131, 803–820 (2016).
Min, H. S., Lee, J. Y., Kim, S. K. & Park, S. H. Genetic grouping of medulloblastomas by representative markers in pathologic diagnosis. Transl Oncol. 6, 265–272 (2013).
Ellison, D. W. et al. Medulloblastoma: clinicopathological correlates of SHH, WNT, and non-SHH/WNT molecular subgroups. Acta Neuropathol. 121, 381–396 (2011).
Northcott, P. A. et al. Rapid, reliable, and reproducible molecular sub-grouping of clinical medulloblastoma samples. Acta Neuropathol. 123, 615–626 (2012).
Leal, L. F. et al. Reproducibility of the NanoString 22-gene molecular subgroup assay for improved prognostic prediction of medulloblastoma. Neuropathology 38, 475–483 (2018).
Schwalbe, E. C. et al. Rapid diagnosis of medulloblastoma molecular subgroups. Clin. Cancer Res. 17, 1883–1894 (2011).
Capper, D. et al. DNA methylation-based classification of central nervous system tumours. Nature 555, 469–474 (2018).
Hovestadt, V. et al. Robust molecular subgrouping and copy-number profiling of medulloblastoma from small amounts of archival tumour material using high-density DNA methylation arrays. Acta Neuropathol. 125, 913–916 (2013).
Schwalbe, E. C. et al. DNA methylation profiling of medulloblastoma allows robust subclassification and improved outcome prediction using formalin-fixed biopsies. Acta Neuropathol. 125, 359–371 (2013). Hovestadt et al. (2013) and Schwalbe et al. (2013) establish the use of genome-wide DNA methylation arrays for molecular subgrouping of MB.
Schwalbe, E. C. et al. Minimal methylation classifier (MIMIC): a novel method for derivation and rapid diagnostic detection of disease-associated DNA methylation signatures. Sci. Rep. 7, 13421 (2017).
Korshunov, A. et al. DNA-methylation profiling discloses significant advantages over NanoString method for molecular classification of medulloblastoma. Acta Neuropathol. 134, 965–967 (2017).
Cavalli, F. M. G. et al. Intertumoral heterogeneity within medulloblastoma subgroups. Cancer Cell 31, 737–754.e6 (2017).
Northcott, P. A. et al. The whole-genome landscape of medulloblastoma subtypes. Nature 547, 311–317 (2017). This study has integrated and expanded earlier next-generation sequencing studies, for the most exhaustive genomic characterization of MB to date.
Schwalbe, E. C. et al. Novel molecular subgroups for clinical classification and outcome prediction in childhood medulloblastoma: a cohort study. Lancet Oncol. 18, 958–971 (2017). Cavalli et al. (2017), Northcott et al. (2017) and Schwalbe et al. (2017) have described further subdivisions of molecular subgroups of MB into subtypes through molecular profiling of increasingly larger patient cohorts.
Kool, M. et al. Molecular subgroups of medulloblastoma: an international meta-analysis of transcriptome, genetic aberrations, and clinical data of WNT, SHH, Group 3, and Group 4 medulloblastomas. Acta Neuropathol. 123, 473–484 (2012).
Gajjar, A. et al. Risk-adapted craniospinal radiotherapy followed by high-dose chemotherapy and stem-cell rescue in children with newly diagnosed medulloblastoma (St Jude Medulloblastoma-96): long-term results from a prospective, multicentre trial. Lancet Oncol. 7, 813–820 (2006).
Ellison, D. W. et al. beta-Catenin status predicts a favorable outcome in childhood medulloblastoma: the United Kingdom Children’s Cancer Study Group Brain Tumour Committee. J. Clin. Oncol. 23, 7951–7957 (2005).
Shih, D. J. et al. Cytogenetic prognostication within medulloblastoma subgroups. J. Clin. Oncol. 32, 886–896 (2014).
Waszak, S. M. et al. Spectrum and prevalence of genetic predisposition in medulloblastoma: a retrospective genetic study and prospective validation in a clinical trial cohort. Lancet Oncol. 19, 785–798 (2018). This study describes germline predisposition to MB, concluding that pathogenic germline mutations account for at least 6% of MB diagnoses.
Remke, M. et al. Adult medulloblastoma comprises three major molecular variants. J. Clin. Oncol. 29, 2717–2723 (2011).
Kool, M. et al. Genome sequencing of SHH medulloblastoma predicts genotype-related response to smoothened inhibition. Cancer Cell 25, 393–405 (2014).
Northcott, P. A. et al. Pediatric and adult sonic hedgehog medulloblastomas are clinically and molecularly distinct. Acta Neuropathol. 122, 231–240 (2011).
Remke, M. et al. TERT promoter mutations are highly recurrent in SHH subgroup medulloblastoma. Acta Neuropathol. 126, 917–929 (2013).
Koelsche, C. et al. Distribution of TERT promoter mutations in pediatric and adult tumors of the nervous system. Acta Neuropathol. 126, 907–915 (2013).
Lindsey, J. C. et al. TERT promoter mutation and aberrant hypermethylation are associated with elevated expression in medulloblastoma and characterise the majority of non-infant SHH subgroup tumours. Acta Neuropathol. 127, 307–309 (2014).
Poschl, J. et al. Genomic and transcriptomic analyses match medulloblastoma mouse models to their human counterparts. Acta Neuropathol. 128, 123–136 (2014).
Robinson, G. W. et al. Risk-adapted therapy for young children with medulloblastoma (SJYC07): therapeutic and molecular outcomes from a multicentre, phase 2 trial. Lancet Oncol. 19, 768–784 (2018). This study defines the molecular landscape of infant MB and integrates molecular findings with a prospective clinical trial cohort, concluding that infant SHH MB can be discriminated into two molecularly distinct subtypes with disparate survival outcomes.
Wang, J., Garancher, A., Ramaswamy, V. & Wechsler-Reya, R. J. Medulloblastoma: from molecular subgroups to molecular targeted therapies. Annu. Rev. Neurosci. 41, 207–232 (2018).
Ramaswamy, V. & Taylor, M. D. Medulloblastoma: from myth to molecular. J. Clin. Oncol. 35, 2355–2363 (2017).
Ramaswamy, V. et al. Risk stratification of childhood medulloblastoma in the molecular era: the current consensus. Acta Neuropathol. 131, 821–831 (2016).
Sharma, T. et al. Second-generation molecular subgrouping of medulloblastoma: an international meta-analysis of Group 3 and Group 4 subtypes. Acta Neuropathol. 138, 309–326 (2019).
Robinson, G. et al. Novel mutations target distinct subgroups of medulloblastoma. Nature 488, 43–48 (2012).
Parsons, D. W. et al. The genetic landscape of the childhood cancer medulloblastoma. Science 331, 435–439 (2011). This is the first exome-level sequencing study of MB, identifying novel recurrent mutations in the chromatin-modifying genes MLL2 and MLL3.
Canning, P. et al. Structural basis for Cul3 protein assembly with the BTB–Kelch family of E3 ubiquitin ligases. J. Biol. Chem. 288, 7803–7814 (2013).
Lee, J. C. et al. Recurrent KBTBD4 small in-frame insertions and absence of DROSHA deletion or DICER1 mutation differentiate pineal parenchymal tumor of intermediate differentiation (PPTID) from pineoblastoma. Acta Neuropathol. 137, 851–854 (2019).
Northcott, P. A. et al. Enhancer hijacking activates GFI1 family oncogenes in medulloblastoma. Nature 511, 428–434 (2014). This study identifies recurrent structural variations leading to the activation of GFI1 and GFI1B in Group 3 MB, without affecting the protein sequence of these oncoproteins.
Moroy, T., Vassen, L., Wilkes, B. & Khandanpour, C. From cytopenia to leukemia: the role of Gfi1 and Gfi1b in blood formation. Blood 126, 2561–2569 (2015).
Erikson, J. et al. Translocation of an immunoglobulin kappa locus to a region 3′ of an unrearranged c-myc oncogene enhances c-myc transcription. Proc. Natl Acad. Sci. USA 80, 7581–7585 (1983).
Croce, C. M. et al. Transcriptional activation of an unrearranged and untranslocated c-myc oncogene by translocation of a C lambda locus in Burkitt. Proc. Natl Acad. Sci. USA 80, 6922–6926 (1983).
Haller, F. et al. Enhancer hijacking activates oncogenic transcription factor NR4A3 in acinic cell carcinomas of the salivary glands. Nat. Commun. 10, 368 (2019).
Martin-Garcia, D. et al. CCND2 and CCND3 hijack immunoglobulin light-chain enhancers in cyclin D1(-) mantle cell lymphoma. Blood 133, 940–951 (2019).
Zimmerman, M. W. et al. MYC drives a subset of high-risk pediatric neuroblastomas and is activated through mechanisms including enhancer hijacking and focal enhancer amplification. Cancer Discov. 8, 320–335 (2018).
Weischenfeldt, J. et al. Pan-cancer analysis of somatic copy-number alterations implicates IRS4 and IGF2 in enhancer hijacking. Nat. Genet. 49, 65–74 (2017).
Ryan, R. J. et al. Detection of enhancer-associated rearrangements reveals mechanisms of oncogene dysregulation in B-cell lymphoma. Cancer Discov. 5, 1058–1071 (2015).
Wu, Y. et al. PRDM6 is enriched in vascular precursors during development and inhibits endothelial cell proliferation, survival, and differentiation. J. Mol. Cell. Cardiol. 44, 47–58 (2008).
Davis, C. A. et al. PRISM/PRDM6, a transcriptional repressor that promotes the proliferative gene program in smooth muscle cells. Mol. Cell. Biol. 26, 2626–2636 (2006).
Hunger, S. P. & Mullighan, C. G. Acute lymphoblastic leukemia in children. N. Engl. J. Med. 373, 1541–1552 (2015).
Parker, M. et al. C11orf95-RELA fusions drive oncogenic NF-κB signalling in ependymoma. Nature 506, 451–455 (2014).
Jones, D. T. et al. Tandem duplication producing a novel oncogenic BRAF fusion gene defines the majority of pilocytic astrocytomas. Cancer Res. 68, 8673–8677 (2008).
He, F. et al. Long noncoding RNA PVT1-214 promotes proliferation and invasion of colorectal cancer by stabilizing Lin28 and interacting with miR-128. Oncogene 38, 164–179 (2019).
Liu, Z. & Zhang, H. LncRNA plasmacytoma variant translocation 1 is an oncogene in bladder urothelial carcinoma. Oncotarget 8, 64273–64282 (2017).
Tian, Z. et al. LncRNA PVT1 regulates growth, migration, and invasion of bladder cancer by miR-31/ CDK1. J. Cell. Physiol. 234, 4799–4811 (2019).
Tseng, Y. Y. et al. PVT1 dependence in cancer with MYC copy-number increase. Nature 512, 82–86 (2014).
Wang, Z., Su, M., Xiang, B., Zhao, K. & Qin, B. Circular RNA PVT1 promotes metastasis via miR-145 sponging in CRC. Biochem. Biophys. Res. Commun. 512, 716–722 (2019).
Yang, T. et al. lncRNA PVT1 and its splicing variant function as competing endogenous RNA to regulate clear cell renal cell carcinoma progression. Oncotarget 8, 85353–85367 (2017).
Zhao, J. et al. LncRNA PVT1 promotes angiogenesis via activating the STAT3/VEGFA axis in gastric cancer. Oncogene 37, 4094–4109 (2018).
Cho, S. W. et al. Promoter of lncRNA gene PVT1 is a tumor-suppressor DNA boundary element. Cell 173, 1398–1412.e22 (2018).
Hovestadt, V. et al. Decoding the regulatory landscape of medulloblastoma using DNA methylation sequencing. Nature 510, 537–541 (2014).
Balzeau, J., Menezes, M. R., Cao, S. & Hagan, J. P. The LIN28/let-7 pathway in cancer. Front. Genet. 8, 31 (2017).
Northcott, P. A. et al. The miR-17/92 polycistron is up-regulated in sonic hedgehog-driven medulloblastomas and induced by N-myc in sonic hedgehog-treated cerebellar neural precursors. Cancer Res. 69, 3249–3255 (2009).
Ferretti, E. et al. MicroRNA profiling in human medulloblastoma. Int. J. Cancer 124, 568–577 (2009).
Uziel, T. et al. The miR-17~92 cluster collaborates with the Sonic Hedgehog pathway in medulloblastoma. Proc. Natl Acad. Sci. USA 106, 2812–2817 (2009).
Zindy, F. et al. Role of the miR-17 approximately 92 cluster family in cerebellar and medulloblastoma development. Biol. Open 3, 597–605 (2014).
Murphy, B. L. et al. Silencing of the miR-17~92 cluster family inhibits medulloblastoma progression. Cancer Res. 73, 7068–7078 (2013).
Bai, A. H. et al. MicroRNA-182 promotes leptomeningeal spread of non-sonic hedgehog-medulloblastoma. Acta Neuropathol. 123, 529–538 (2012).
Weeraratne, S. D. et al. Pleiotropic effects of miR-183~96~182 converge to regulate cell survival, proliferation and migration in medulloblastoma. Acta Neuropathol. 123, 539–552 (2012).
Zhang, Z., Li, S. & Cheng, S. Y. The miR-183 approximately 96 approximately 182 cluster promotes tumorigenesis in a mouse model of medulloblastoma. J. Biomed. Res. 27, 486–494 (2013).
Ferretti, E. et al. Concerted microRNA control of Hedgehog signalling in cerebellar neuronal progenitor and tumour cells. EMBO J. 27, 2616–2627 (2008).
Su, X. et al. Abnormal expression of REST/NRSF and Myc in neural stem/progenitor cells causes cerebellar tumors by blocking neuronal differentiation. Mol. Cell. Biol. 26, 1666–1678 (2006).
Wu, J. & Xie, X. Comparative sequence analysis reveals an intricate network among REST, CREB and miRNA in mediating neuronal gene expression. Genome Biol. 7, R85 (2006).
Pierson, J., Hostager, B., Fan, R. & Vibhakar, R. Regulation of cyclin dependent kinase 6 by microRNA 124 in medulloblastoma. J. Neurooncol. 90, 1–7 (2008).
Silber, J. et al. Expression of miR-124 inhibits growth of medulloblastoma cells. Neuro-Oncology 15, 83–90 (2013).
Wang, X. et al. miR miR on the wall, who’s the most malignant medulloblastoma miR of them all? Neuro-Oncology 20, 313–323 (2018).
Hansen, K. D. et al. Increased methylation variation in epigenetic domains across cancer types. Nat. Genet. 43, 768–775 (2011).
Berman, B. P. et al. Regions of focal DNA hypermethylation and long-range hypomethylation in colorectal cancer coincide with nuclear lamina-associated domains. Nat. Genet. 44, 40–46 (2011).
Hon, G. C. et al. Global DNA hypomethylation coupled to repressive chromatin domain formation and gene silencing in breast cancer. Genome Res. 22, 246–258 (2012).
Lin, C. Y. et al. Active medulloblastoma enhancers reveal subgroup-specific cellular origins. Nature 530, 57–62 (2016).
Consortium, E. P. An integrated encyclopedia of DNA elements in the human genome. Nature 489, 57–74 (2012).
Thurman, R. E. et al. The accessible chromatin landscape of the human genome. Nature 489, 75–82 (2012).
Roadmap Epigenomics Consortium et al. Integrative analysis of 111 reference human epigenomes. Nature 518, 317–330 (2015).
Aebersold, R. & Mann, M. Mass-spectrometric exploration of proteome structure and function. Nature 537, 347–355 (2016).
Archer, T. C. et al. Proteomics, post-translational modifications, and integrative analyses reveal molecular heterogeneity within medulloblastoma subgroups. Cancer Cell 34, 396–410 e398 (2018).
Forget, A. et al. Aberrant ERBB4-SRC signaling as a hallmark of group 4 medulloblastoma revealed by integrative phosphoproteomic profiling. Cancer Cell 34, 379–395.e377 (2018). Archer et al. (2018) and Forget et al. (2018) first describe quantification of the global proteome and phospho-proteome landscape in primary patient samples through mass spectrometry.
Rivero-Hinojosa, S. et al. Proteomic analysis of medulloblastoma reveals functional biology with translational potential. Acta Neuropathol. Commun. 6, 48 (2018).
Vogel, C. & Marcotte, E. M. Insights into the regulation of protein abundance from proteomic and transcriptomic analyses. Nat. Rev. Genet. 13, 227–232 (2012).
Staal, J. A. et al. Proteomic profiling of high risk medulloblastoma reveals functional biology. Oncotarget 6, 14584–14595 (2015).
Zomerman, W. W. et al. Identification of two protein-signaling states delineating transcriptionally heterogeneous human medulloblastoma. Cell Rep. 22, 3206–3216 (2018).
Canettieri, G. et al. Histone deacetylase and Cullin3-REN(KCTD11) ubiquitin ligase interplay regulates Hedgehog signalling through Gli acetylation. Nat. Cell Biol. 12, 132–142 (2010).
Klisch, T. J., Vainshtein, A., Patel, A. J. & Zoghbi, H. Y. Jak2-mediated phosphorylation of Atoh1 is critical for medulloblastoma growth. eLife 6, 31181 (2017).
Al-Hajj, M., Wicha, M. S., Benito-Hernandez, A., Morrison, S. J. & Clarke, M. F. Prospective identification of tumorigenic breast cancer cells. Proc. Natl Acad. Sci. USA 100, 3983–3988 (2003).
Lapidot, T. et al. A cell initiating human acute myeloid leukaemia after transplantation into SCID mice. Nature 367, 645–648 (1994).
Bonnet, D. & Dick, J. E. Human acute myeloid leukemia is organized as a hierarchy that originates from a primitive hematopoietic cell. Nat. Med. 3, 730–737 (1997).
Singh, S. K. et al. Identification of a cancer stem cell in human brain tumors. Cancer Res. 63, 5821–5828 (2003).
Singh, S. K. et al. Identification of human brain tumour initiating cells. Nature 432, 396–401 (2004). This study first reports the discovery of a CD133+ stem-like tumour cell population in MB that can efficiently initiate tumours in vivo, whereas the remaining CD133− tumour cells cannot.
Garg, N. et al. CD133+ brain tumor-initiating cells are dependent on STAT3 signaling to drive medulloblastoma recurrence. Oncogene 36, 606–617 (2017).
Read, T. A. et al. Identification of CD15 as a marker for tumor-propagating cells in a mouse model of medulloblastoma. Cancer Cell 15, 135–147 (2009).
Ward, R. J. et al. Multipotent CD15 + cancer stem cells in patched-1-deficient mouse medulloblastoma. Cancer Res. 69, 4682–4690 (2009).
Vanner, R. J. et al. Quiescent sox2+ cells drive hierarchical growth and relapse in sonic hedgehog subgroup medulloblastoma. Cancer Cell 26, 33–47 (2014).
Weng, Q. et al. Single-cell transcriptomics uncovers glial progenitor diversity and cell fate determinants during development and gliomagenesis. Cell Stem Cell 24, 707–723.e708 (2019).
Filbin, M. G. et al. Developmental and oncogenic programs in H3K27M gliomas dissected by single-cell RNA-seq. Science 360, 331–335 (2018).
Venteicher, A. S. et al. Decoupling genetics, lineages, and microenvironment in IDH-mutant gliomas by single-cell RNA-seq. Science 355, eaai8478 (2017).
Tirosh, I. et al. Single-cell RNA-seq supports a developmental hierarchy in human oligodendroglioma. Nature 539, 309–313 (2016).
Patel, A. P. et al. Single-cell RNA-seq highlights intratumoral heterogeneity in primary glioblastoma. Science 344, 1396–1401 (2014).
Vladoiu, M. C. et al. Childhood cerebellar tumours mirror conserved fetal transcriptional programs. Nature 572, 67–73 (2019).
Hovestadt, V. et al. Resolving medulloblastoma cellular architecture by single-cell genomics. Nature 572, 74–79 (2019). Vladoiu et al. (2019) and Hovestadt et al. (2019) first apply single-cell transcriptome profiling to characterizing cellular heterogeneity within individual patients with MB and identifying developmental correlates from the developing mouse cerebellum.
Oliver, T. G. et al. Loss of patched and disruption of granule cell development in a pre-neoplastic stage of medulloblastoma. Development 132, 2425–2439 (2005).
Morrissy, A. S. et al. Spatial heterogeneity in medulloblastoma. Nat. Genet. 49, 780–788 (2017).
Fults, D. W., Taylor, M. D. & Garzia, L. Leptomeningeal dissemination: a sinister pattern of medulloblastoma growth. J. Neurosurg. https://doi.org/10.3171/2018.11.PEDS18506 (2019).
Ramaswamy, V. et al. Recurrence patterns across medulloblastoma subgroups: an integrated clinical and molecular analysis. Lancet Oncol. 14, 1200–1207 (2013).
Hill, R. M. et al. Combined MYC and P53 defects emerge at medulloblastoma relapse and define rapidly progressive, therapeutically targetable disease. Cancer Cell 27, 72–84 (2015).
Morrissy, A. S. et al. Divergent clonal selection dominates medulloblastoma at recurrence. Nature 529, 351–357 (2016).
Zapotocky, M. et al. Differential patterns of metastatic dissemination across medulloblastoma subgroups. J. Neurosurg. 21, 145–152 (2018).
Garzia, L. et al. A hematogenous route for medulloblastoma leptomeningeal metastases. Cell 173, 1549 (2018). This study describes the presence of circulating tumour cells in the blood of patients that can spread to form leptomeningeal metastases.
US National Library of Medicine. ClinicalTrials.gov https://clinicaltrials.gov/ct2/show/NCT01878617 (2013).
US National Library of Medicine. ClinicalTrials.gov https://clinicaltrials.gov/ct2/show/NCT02066220 (2014).
US National Library of Medicine. ClinicalTrials.gov https://clinicaltrials.gov/ct2/show/NCT02724579 (2016).
Sharma, T. et al. Second-generation molecular subgrouping of medulloblastoma: an international meta-analysis of Group 3 and Group 4 subtypes. Acta Neuropathol. 138, 309–326 (2019).
Druker, H. et al. Genetic counselor recommendations for cancer predisposition evaluation and surveillance in the pediatric oncology patient. Clin. Cancer Res. 23, e91–e97 (2017).
Romer, J. T. et al. Suppression of the Shh pathway using a small molecule inhibitor eliminates medulloblastoma in Ptc1+/–p53–/– mice. Cancer Cell 6, 229–240 (2004).
Lee, Y. et al. Loss of suppressor-of-fused function promotes tumorigenesis. Oncogene 26, 6442–6447 (2007).
Robinson, G. W. et al. Vismodegib exerts targeted efficacy against recurrent sonic hedgehog-subgroup medulloblastoma: results from phase II Pediatric Brain Tumor Consortium studies PBTC-025B and PBTC-032. J. Clin. Oncol. 33, 2646–2654 (2015).
Robinson, G. W. et al. Irreversible growth plate fusions in children with medulloblastoma treated with a targeted hedgehog pathway inhibitor. Oncotarget 8, 69295–69302 (2017).
Kimura, H., Ng, J. M. & Curran, T. Transient inhibition of the Hedgehog pathway in young mice causes permanent defects in bone structure. Cancer Cell 13, 249–260 (2008).
Lee, C. et al. Lsd1 as a therapeutic target in Gfi1-activated medulloblastoma. Nat. Commun. 10, 332 (2019).
Morabito, M. et al. An autocrine ActivinB mechanism drives TGFβ/activin signaling in Group 3 medulloblastoma. EMBO Mol. Med. 11, e9830 (2019).
Gilbertson, R. J. & Ellison, D. W. The origins of medulloblastoma subtypes. Annu. Rev. Pathol. 3, 341–365 (2008).
Gibson, P. et al. Subtypes of medulloblastoma have distinct developmental origins. Nature 468, 1095–1099 (2010).
Swartling, F. J. et al. Distinct neural stem cell populations give rise to disparate brain tumors in response to N-MYC. Cancer Cell 21, 601–613 (2012).
Pei, Y. et al. An animal model of MYC-driven medulloblastoma. Cancer Cell 21, 155–167 (2012).
Kawauchi, D. et al. Novel MYC-driven medulloblastoma models from multiple embryonic cerebellar cells. Oncogene 36, 5231–5242 (2017).
Kawauchi, D. et al. A mouse model of the most aggressive subgroup of human medulloblastoma. Cancer Cell 21, 168–180 (2012).
Swartling, F. J. et al. Pleiotropic role for MYCN in medulloblastoma. Genes Dev. 24, 1059–1072 (2010).
Chizhikov, V. V. et al. Lmx1a regulates fates and location of cells originating from the cerebellar rhombic lip and telencephalic cortical hem. Proc. Natl Acad. Sci. USA 107, 10725–10730 (2010).
Englund, C. et al. Unipolar brush cells of the cerebellum are produced in the rhombic lip and migrate through developing white matter. J. Neurosci. 26, 9184–9195 (2006).
Acknowledgements
P.A.N. is a Pew–Stewart Scholar for Cancer Research (Margaret and Alexander Stewart Trust) and recipient of the Sontag Foundation Distinguished Scientist Award. P.A.N. was also supported by the National Cancer Institute (R01CA232143-01), the American Association for Cancer Research (NextGen Grant for Transformative Cancer Research), The Brain Tumour Charity (Quest for Cures and Clinical Biomarkers), the American Lebanese Syrian Associated Charities (ALSAC) and St. Jude. V.H. is supported by a Human Frontier Science Program long-term fellowship (LT000596/2016-L). We acknowledge K. Smith, L. Bihannic and T. Sharma for assistance with data organization and figure design. We thank B. Stelter for assistance with the artwork.
Author information
Authors and Affiliations
Contributions
V.H. and P.A.N researched data for the article, contributed substantially to discussion of the content, wrote the article and reviewed/edited the manuscript before submission. O.A., F.J.S., G.W.R. and S.M.P. researched data for the article, wrote the article and reviewed/edited the manuscript before submission.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information
Nature Reviews Cancer thanks T. Curran, E. Ferretti and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Glossary
- Next-generation sequencing
-
(NGS). Technologies enabling massively parallel reading of amplified short nucleotide sequences (typically yielding hundreds of millions of reads, 100–500 bp in length). In contrast, emerging third-generation sequencing technologies read sequences without prior amplification, yielding much longer reads, albeit with reduced accuracy and throughput.
- Molecular classification
-
Classification of patient tumour samples based on molecular markers, as opposed to classification based on histomorphological appearance.
- Intratumoural heterogeneity
-
The observation that tumours comprise distinct malignant and non-malignant (cells of the microenvironment) cell types. The heterogeneity of malignant cells encompasses genetic heterogeneity (for example, different genetic subclones) and transcriptional heterogeneity (for example, malignant cell states resembling normal development).
- Primitive neuroectodermal tumour
-
(PNET). A class of histologically defined, poorly differentiated childhood brain tumours. More recently, PNET has been reclassified into a number of both novel and previously known brain tumour entities by molecular profiling.
- Atypical teratoid rhabdoid tumour
-
A rare and highly malignant type of childhood brain tumour that is characterized by mutations in the SMARCB1 gene.
- Classic histology
-
The most common histological variant found in all medulloblastoma subgroups, characterized by densely packed small round or oval cells and a high nuclear:cytoplasmic ratio.
- Desmoplastic/nodular histology
-
A histological variant mostly restricted to Sonic hedgehog medulloblastoma, characterized by a varying number of islands of neurocytic differentiation and internodular desmoplasia.
- Large-cell/anaplastic (LCA) histology
-
A histological variant associated with both Group 3 medulloblastoma and a more aggressive phenotype, characterized by the co-occurrence of groups of large cells with round nuclei and cells exhibiting cytological pleomorphism (anaplasia).
- Isochromosome
-
An abnormal chromosome in which both chromosome arms are identical. Isochromosome 17q is one of the most frequent somatic copy-number alterations in Group 3 and Group 4 medulloblastoma. In most cases a second q-arm is fused to the p-arm proximal to the centromere.
- Craniospinal axis irradiation
-
(CSI). Standard therapy for patients with medulloblastoma following surgery, to reduce risk of tumour regrowth and metastatic dissemination. The application of CSI is also associated with neurological impairments and secondary malignancies.
- Non-synonymous mutations
-
Genetic alterations that alter the amino acid sequence of an affected protein, possibly altering protein function. Most described recurrent mutations in medulloblastoma are non-synonymous mutations.
- BTB–Kelch protein family
-
Family of proteins characterized by the presence of an N-terminal BTB domain and C-terminal Kelch motifs. The BTB domain facilitates protein binding, and the Kelch motifs associate to form a β-propeller facilitating protein–protein interactions. This family function as adaptor proteins that link cullin–RING ligases to substrates for ubiquitylation.
- Pineal parenchymal tumours of intermediate differentiation
-
(PPTID). A very rare tumour type of intermediate grade arising from the pineal parenchyma.
- Chromothripsis
-
Clustered occurrence of a large number of structural variants restricted to a single chromosome or chromosomal arm, emerging through a single catastrophic event.
- Ptch1+/– mouse MB model
-
Transgenic mouse model that is heterozygous for the Ptch1 gene. Sporadic Sonic hedgehog medulloblastoma develops in ~15% of Ptch1+/– mice.
- Single-cell RNA sequencing
-
Emerging technology that enables unsupervised characterization of transcriptional profiles in individual cells of healthy and diseased tissues. The throughput of this technology has steadily increased over recent years, now enabling profiling of tens of thousands of individual cells in a single experiment.
- Granule neuron progenitor
-
(GNP). A progenitor cell type that gives rise to granule cells, the most common type of neuron in the mature cerebellum; the presumed developmental origin of Sonic hedgehog medulloblastoma.
- Unipolar brush cells
-
(UBCs). Rare glutamatergic interneurons found in the cerebellar cortex and in the dorsal cochlear nucleus. Recent studies have identified transcriptional similarities between UBCs in mouse and human Group 4 medulloblastoma.
- Glutamatergic cerebellar nuclei
-
(GluCN). Also referred to as deep cerebellar nuclei. Cells that function (along with GABAergic interneurons) as the main output centres of the cerebellum.
- Sleeping Beauty transposon system
-
A synthetic DNA transposon system used for random mutagenesis screening and in a recent Sonic hedgehog-medulloblastoma mouse model. Genes affected by genomic insertion of the transposon can be identified through sequencing.
- Bone age
-
Degree of skeletal maturity, an important parameter for determining the clinical use of SMO inhibitors in patients with Sonic hedgehog medulloblastoma. Prolonged exposure to the targeted inhibitor vismodegib has been associated with growth defects in children that have not reached skeletal maturity.
- Patient-derived xenograft
-
(PDX). A model of cancer in which tumour cells from a patient are implanted and maintained in a non-human carrier, most commonly immunodeficient or humanized laboratory mice. PDX models are thought to resemble patient tumours more closely than cell cultures do.
- Blood–brain barrier
-
A semipermeable border formed by endothelial cells lining the cerebral microvasculature that separates the brain from the circulating blood and protects the brain from fluctuations in plasma composition and from circulating agents such as neurotransmitters and pathogens. The blood–brain barrier also presents a challenge for drug delivery when treating brain tumours.
- Blood–tumour barrier
-
Tumour-associated compromise of the blood–brain barrier, resulting in a highly heterogeneous vasculature characterized by non-uniform permeability to small and large molecules.
Rights and permissions
About this article
Cite this article
Hovestadt, V., Ayrault, O., Swartling, F.J. et al. Medulloblastomics revisited: biological and clinical insights from thousands of patients. Nat Rev Cancer 20, 42–56 (2020). https://doi.org/10.1038/s41568-019-0223-8
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41568-019-0223-8
This article is cited by
-
Marinopyrrole derivative MP1 as a novel anti-cancer agent in group 3 MYC-amplified Medulloblastoma
Journal of Experimental & Clinical Cancer Research (2024)
-
SALL4 is a CRL3REN/KCTD11 substrate that drives Sonic Hedgehog-dependent medulloblastoma
Cell Death & Differentiation (2024)
-
Evolution of neurosurgical advances and nuances in medulloblastoma therapy
Child's Nervous System (2024)
-
Compartments in medulloblastoma with extensive nodularity are connected through differentiation along the granular precursor lineage
Nature Communications (2024)
-
Mitochondrial DNA mutations in Medulloblastoma
Acta Neuropathologica Communications (2023)