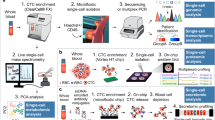
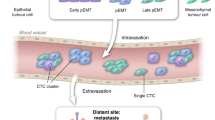
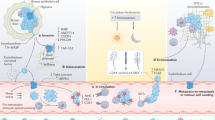
Abstract
Single-cell technologies have contributed to unravelling tumour heterogeneity, now considered a hallmark of cancer and one of the main causes of tumour resistance to cancer therapies. Liquid biopsy (LB), defined as the detection and analysis of cells or cell products released by tumours into the blood, offers an appealing minimally invasive approach that allows the characterization and monitoring of tumour heterogeneity in individual patients. Here, we will review and discuss how circulating tumour cell (CTC) analysis at single-cell resolution provides unique insights into tumour heterogeneity that are not revealed by analysis of circulating tumour DNA (ctDNA) derived from LBs. The molecular analysis of CTCs provides complementary information to that of genomic aberrations determined using ctDNA to fully describe many different cellular components (for example, DNA, RNA, proteins and metabolites) that can influence tumour heterogeneity.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
References
Alizadeh, A. A. et al. Toward understanding and exploiting tumor heterogeneity. Nat. Med. 21, 846–853 (2015).
McGranahan, N. & Swanton, C. Clonal heterogeneity and tumor evolution: past, present, and the future. Cell 168, 613–628 (2017).
Lindstrom, L. S. et al. Clinically used breast cancer markers such as estrogen receptor, progesterone receptor, and human epidermal growth factor receptor 2 are unstable throughout tumor progression. J. Clin. Oncol. 30, 2601–2608 (2012).
Gerlinger, M. et al. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N. Engl. J. Med. 366, 883–892 (2012).
Pantel, K. & Alix-Panabieres, C. Circulating tumour cells in cancer patients: challenges and perspectives. Trends Mol. Med. 16, 398–406 (2010).
Mostert, B. et al. Gene expression profiles in circulating tumor cells to predict prognosis in metastatic breast cancer patients. Ann. Oncol. 26, 510–516 (2014).
Chimonidou, M., Strati, A., Malamos, N., Georgoulias, V. & Lianidou, E. S. SOX17 promoter methylation in circulating tumor cells and matched cell-free DNA isolated from plasma of patients with breast cancer. Clin. Chem. 59, 270–279 (2013).
Heitzer, E., Haque, I. S., Roberts, C. E. S. & Speicher, M. R. Current and future perspectives of liquid biopsies in genomics-driven oncology. Nat. Rev. Genet. 20, 71–88 (2019).
Anfossi, S., Babayan, A., Pantel, K. & Calin, G. A. Clinical utility of circulating non-coding RNAs—an update. Nat. Rev. Clin. Oncol. 15, 541–563 (2018).
Xu, R. et al. Extracellular vesicles in cancer—implications for future improvements in cancer care. Nat. Rev. Clin. Oncol. 15, 617–638 (2018).
Best, M. G. et al. RNA-Seq of tumor-educated platelets enables blood-based pan-cancer, multiclass, and molecular pathway cancer diagnostics. Cancer Cell 28, 666–676 (2015).
Leong, S. P. & Tseng, W. W. Micrometastatic cancer cells in lymph nodes, bone marrow, and blood: clinical significance and biologic implications. CA Cancer J. Clin. 64, 195–206 (2014).
Chiang, S. P. H., Cabrera, R. M. & Segall, J. E. Tumor cell intravasation. Am. J. Physiol. Cell Physiol. 311,C1–C14 (2016).
Harney, A. S. et al. Real-time imaging reveals local, transient vascular permeability, and tumor cell intravasation stimulated by TIE2hi macrophage-derived VEGFA. Cancer Discov. 5, 932–943 (2015).
Brown, M. et al. Lymph node blood vessels provide exit routes for metastatic tumor cell dissemination in mice. Science 359, 1408–1411 (2018).
Pereira, E. R. et al. Lymph node metastases can invade local blood vessels, exit the node, and colonize distant organs in mice. Science 359, 1403–1407 (2018). This paper and that by Brown et al. (2018) demonstrate that tumour cells can reach peripheral blood circulation through invasion into blood vessels within lymph nodes.
Naxerova, K. et al. Origins of lymphatic and distant metastases in human colorectal cancer. Science 357, 55–60 (2017).
Rack, B. et al. Circulating tumor cells predict survival in early average-to-high risk breast cancer patients. J. Natl Cancer Inst. 106, dju066 (2014).
Goodman, C. R. et al. Association of circulating tumor cell status with benefit of radiotherapy and survival in early-stage breast cancer. JAMA Oncol. 4, e180163 (2018).
Trapp, E. et al. Presence of circulating tumor cells in high-risk early breast cancer during follow-up and prognosis. J. Natl Cancer Inst. 111, 380–387 (2019).
Kim, M. Y. et al. Tumor self-seeding by circulating cancer cells. Cell 139, 1315–1326 (2009).
Garzia, L. et al. A hematogenous route for medulloblastoma leptomeningeal metastases. Cell 173, 1549 (2018).
Pradeep, S. et al. Hematogenous metastasis of ovarian cancer: rethinking mode of spread. Cancer Cell 26, 77–91 (2014).
Massague, J. & Obenauf, A. C. Metastatic colonization by circulating tumour cells. Nature 529, 298–306 (2016).
Lyle, L. T. et al. Alterations in pericyte subpopulations are associated with elevated blood-tumor barrier permeability in experimental brain metastasis of breast cancer. Clin. Cancer Res. 22, 5287–5299 (2016).
Wan, J. C. M. et al. Liquid biopsies come of age: towards implementation of circulating tumour DNA. Nat. Rev. Cancer 17, 223 (2017).
Hanssen, A. et al. Frequency of circulating tumor cells (CTC) in patients with brain metastases: implications as a risk assessment marker in oligo-metastatic disease. Cancers 10, E52 (2018).
Muller, C. et al. Hematogenous dissemination of glioblastoma multiforme. Sci. Transl. Med. 6, 247ra101 (2014).
Sullivan, J. P. et al. Brain tumor cells in circulation are enriched for mesenchymal gene expression. Cancer Discov. 4, 1299–1309 (2014). This paper and that by Muller et al. (2014) challenge the dogma that glioblastoma spread is restricted to the brain by demonstrating that glioma-derived CTCs are also detectable in peripheral blood.
Liu, T. et al. Circulating glioma cells exhibit stem cell-like properties. Cancer Res. 78, 6632–6642 (2018).
Kai, F., Drain, A. P. & Weaver, V. M. The extracellular matrix modulates the metastatic journey. Dev. Cell 49, 332–346 (2019).
Te Boekhorst, V. & Friedl, P. Plasticity of cancer cell invasion-mechanisms and implications for therapy. Adv. Cancer Res. 132, 209–264 (2016).
Friedl, P. Prespecification and plasticity: shifting mechanisms of cell migration. Curr. Opin. Cell Biol. 16, 14–23 (2004).
Georgouli, M. et al. Regional activation of myosin II in cancer cells drives tumor progression via a secretory cross-talk with the immune microenvironment. Cell 176, 757–774.e723 (2019).
Rodriguez-Hernandez, I., Cantelli, G., Bruce, F. & Sanz-Moreno, V. Rho, ROCK and actomyosin contractility in metastasis as drug targets. F1000Research 5, F1000 (2016).
Cantelli, G. et al. TGF-β-induced transcription sustains amoeboid melanoma migration and dissemination. Curr. Biol. 25, 2899–2914 (2015).
Liu, Y. J. et al. Confinement and low adhesion induce fast amoeboid migration of slow mesenchymal cells. Cell 160, 659–672 (2015).
Taddei, M. L. et al. EphA2 induces metastatic growth regulating amoeboid motility and clonogenic potential in prostate carcinoma cells. Mol. Cancer Res. 9, 149–160 (2011).
Taddei, M. L. et al. Mesenchymal to amoeboid transition is associated with stem-like features of melanoma cells. Cell Commun. Signal. 12, 24 (2014).
Berton, S. et al. The tumor suppressor functions ofp27(kip1) include control of the mesenchymal/amoeboid transition. Mol. Cell. Biol. 29, 5031–5045 (2009).
Aceto, N. et al. Circulating tumor cell clusters are oligoclonal precursors of breast cancer metastasis. Cell 158, 1110–1122 (2014).
Hou, J. M. et al. Clinical significance and molecular characteristics of circulating tumor cells and circulating tumor microemboli in patients with small-cell lung cancer. J. Clin. Oncol. 30, 525–532 (2012).
Andree, K. C., van Dalum, G. & Terstappen, L. W. Challenges in circulating tumor cell detection by the CellSearch system. Mol. Oncol. 10, 395–407 (2016).
Friedl, P., Locker, J., Sahai, E. & Segall, J. E. Classifying collective cancer cell invasion. Nat. Cell Biol. 14, 777 (2012).
Pandya, P., Orgaz, J. L. & Sanz-Moreno, V. Actomyosin contractility and collective migration: may the force be with you. Curr. Opin. Cell Biol. 48, 87–96 (2017).
Liu, X. et al. Homophilic CD44 interactions mediate tumor cell aggregation and polyclonal metastasis in patient-derived breast cancer models. Cancer Discov. 9, 96–113 (2019).
Au, S. H. et al. Clusters of circulating tumor cells traverse capillary-sized vessels. Proc. Natl Acad. Sci. USA 113, 4947–4952 (2016).
Yu, M. et al. Circulating breast tumor cells exhibit dynamic changes in epithelial and mesenchymal composition. Science 339, 580–584 (2013).
Aiello, N. M. et al. EMT subtype influences epithelial plasticity and mode of cell migration. Dev. Cell 45, 681–695.e684 (2018).
Gkountela, S. et al. Circulating tumor cell clustering shapes DNA methylation to enable metastasis seeding. Cell 176, 98–112.e114 (2019). These study results link CTC clustering to specific changes in DNA methylation that promote cancer stemness and metastasis, and point to CTC cluster-targeting drug compounds as a new approach to suppress cancer cell dissemination.
Duda, D. G. et al. Malignant cells facilitate lung metastasis by bringing their own soil. Proc. Natl Acad. Sci. USA 107, 21677–21682 (2010).
Szczerba, B. M. et al. Neutrophils escort circulating tumour cells to enable cell cycle progression. Nature 566, 553–557 (2019).
Sprouse, M. L. et al. PMN-MDSCs enhance CTC metastatic properties through reciprocal interactions via ROS/Notch/Nodal signaling. Int. J. Mol. Sci. 20, E1916 (2019).
Chen, M. B. et al. Inflamed neutrophils sequestered at entrapped tumor cells via chemotactic confinement promote tumor cell extravasation. Proc. Natl Acad. Sci. USA 115, 7022–7027 (2018).
Wang, C. et al. Longitudinally collected CTCs and CTC-clusters and clinical outcomes of metastatic breast cancer. Breast Cancer Res. Treat. 161, 83–94 (2017).
Fanelli, M. F. et al. Evaluation of incidence, significance, and prognostic role of circulating tumor microemboli and transforming growth factor-β receptor I in head and neck cancer. Head Neck 39, 2283–2292 (2017).
Mani, S. A. et al. The epithelial–mesenchymal transition generates cells with properties of stem cells. Cell 133, 704–715 (2008).
Aktas, B. et al. Stem cell and epithelial–mesenchymal transition markers are frequently overexpressed in circulating tumor cells of metastatic breast cancer patients. Breast Cancer Res. 11, R46 (2009).
Papadaki, M. A. et al. Co-expression of putative stemness and epithelial-to-mesenchymal transition markers on single circulating tumour cells from patients with early and metastatic breast cancer. BMC Cancer 14, 651 (2014).
Markiewicz, A. et al. Aggressive phenotype of cells disseminated via hematogenous and lymphatic route in breast cancer patients. Transl. Oncol. 11, 722–731 (2018).
Markiewicz, A. et al. Spectrum of epithelial–mesenchymal transition phenotypes in circulating tumour cells from early breast cancer patients. Cancers 11, E59 (2019).
Cayrefourcq, L. et al. Establishment and characterization of a cell line from human circulating colon cancer cells. Cancer Res. 75, 892–901 (2015).
Hodgkinson, C. L. et al. Tumorigenicity and genetic profiling of circulating tumor cells in small-cell lung cancer. Nat. Med. 20, 897–903 (2014).
Abbosh, C. et al. Phylogenetic ctDNA analysis depicts early-stage lung cancer evolution. Nature 545, 446–451 (2017). This report of the TRACERx study shows how phylogenetic ctDNA profiling can track the subclonal nature of lung cancer relapse and metastasis.
Baccelli, I. et al. Identification of a population of blood circulating tumor cells from breast cancer patients that initiates metastasis in a xenograft assay. Nat. Biotechnol. 31, 539–544 (2013).
Zhang, L. et al. The identification and characterization of breast cancer CTCs competent for brain metastasis. Sci. Transl. Med. 5, 180ra148 (2013).
Boral, D. et al. Molecular characterization of breast cancer CTCs associated with brain metastasis. Nat. Commun. 8, 196 (2017).
Wu, Z. et al. TPO-induced metabolic reprogramming drives liver metastasis of colorectal cancer CD110+ tumor-initiating cells. Cell Stem Cell 17, 47–59 (2015).
Alix-Panabieres, C. et al. Molecular portrait of metastasis-competent circulating tumor cells in colon cancer reveals the crucial role of genes regulating energy metabolism and DNA repair. Clin. Chem. 63, 700–713 (2017).
Vishnoi, M. et al. Targeting USP7 identifies a metastasis-competent state within bone marrow-resident melanoma CTCs. Cancer Res. 78, 5349–5362 (2018).
Girotti, M. R. et al. Application of sequencing, liquid biopsies, and patient-derived xenografts for personalized medicine in melanoma. Cancer Discov. 6, 286–299 (2016).
Follain, G. et al. Hemodynamic forces tune the arrest, adhesion, and extravasation of circulating tumor cells. Dev. Cell 45, 33–52.e12 (2018).
Yano, K. et al. Phenotypic heterogeneity is an evolutionarily conserved feature of the endothelium. Blood 109, 613–615 (2007).
Yasmin-Karim, S., King, M. R., Messing, E. M. & Lee, Y. F. E-selectin ligand-1 controls circulating prostate cancer cell rolling/adhesion and metastasis. Oncotarget 5, 12097–12110 (2014).
Tichet, M. et al. Tumour-derived SPARC drives vascular permeability and extravasation through endothelial VCAM1 signalling to promote metastasis. Nat. Commun. 6, 6993 (2015).
Esposito, M. et al. Bone vascular niche E-selectin induces mesenchymal–epithelial transition and Wnt activation in cancer cells to promote bone metastasis. Nat. Cell Biol. 21, 627–639 (2019). This study reports that bone vascular niches promote bone metastasis by activating cancer stem cell properties and inducing mesenchymal–epithelial transition required for metastatic outgrowth.
Er, E. E. et al. Pericyte-like spreading by disseminated cancer cells activates YAP and MRTF for metastatic colonization. Nat. Cell. Biol. 20, 966–978 (2018).
Amirouchene-Angelozzi, N., Swanton, C. & Bardelli, A. Tumor evolution as a therapeutic target. Cancer Discov. 7, 805–817 (2017).
Swanton, C., McGranahan, N., Starrett, G. J. & Harris, R. S. APOBEC enzymes: mutagenic fuel for cancer evolution and heterogeneity. Cancer Discov. 5, 704–712 (2015).
Mishima, Y. et al. The mutational landscape of circulating tumor cells in multiple myeloma. Cell Rep. 19, 218–224 (2017).
Lohr, J. G. et al. Whole-exome sequencing of circulating tumor cells provides a window into metastatic prostate cancer. Nat. Biotechnol. 32, 479–484 (2014).
Lambros, M. B. et al. Single-cell analyses of prostate cancer liquid biopsies acquired by apheresis. Clin. Cancer Res. 24, 5635–5644 (2018). This report shows that DLA facilitates the capture of very large numbers of CTCs, which allows the deconvolution of intra-patient heterogeneity and clonal evolution.
Paoletti, C. et al. Comprehensive mutation and copy number profiling in archived circulating breast cancer tumor cells documents heterogeneous resistance mechanisms. Cancer Res. 78, 1110–1122 (2018).
Turajlic, S. & Swanton, C. Metastasis as an evolutionary process. Science 352, 169–175 (2016).
Gao, Y. et al. Single-cell sequencing deciphers a convergent evolution of copy number alterations from primary to circulating tumor cells. Genome Res. 27, 1312–1322 (2017).
Joosse, S. A. et al. Chromosomal aberrations associated with sequential steps of the metastatic cascade in colorectal cancer patients. Clin. Chem. 64, 1505–1512 (2018).
Nong, J. et al. Circulating tumor DNA analysis depicts subclonal architecture and genomic evolution of small cell lung cancer. Nat. Commun. 9, 3114 (2018).
Beltran, H. et al. The initial detection and partial characterization of circulating tumor cells in neuroendocrine prostate cancer. Clin. Cancer Res. 22, 1510–1519 (2016).
Chimonidou, M. et al. Direct comparison study of DNA methylation markers in EpCAM-positive circulating tumour cells, corresponding circulating tumour DNA, and paired primary tumours in breast cancer. Oncotarget 8, 72054–72068 (2017).
Shaw, J. A. et al. Mutation analysis of cell-free DNA and single circulating tumor cells in metastatic breast cancer patients with high circulating tumor cell counts. Clin. Cancer Res. 23, 88–96 (2017).
Werner, S. et al. Suppression of early hematogenous dissemination of human breast cancer cells to bone marrow by retinoic acid-induced 2. Cancer Discov. 5, 506–519 (2015).
Wrage, M. et al. Genomic profiles associated with early micrometastasis in lung cancer: relevance of 4q deletion. Clin. Cancer Res. 15, 1566–1574 (2009).
Powell, A. A. et al. Single cell profiling of circulating tumor cells: transcriptional heterogeneity and diversity from breast cancer cell lines. PLOS ONE 7, e33788 (2012).
Gorges, T. M. et al. Accession of tumor heterogeneity by multiplex transcriptome profiling of single circulating tumor cells. Clin. Chem. 62, 1504–1515 (2016).
Ledergor, G. et al. Single cell dissection of plasma cell heterogeneity in symptomatic and asymptomatic myeloma. Nat. Med. 24, 1867–1876 (2018).
Loomans-Kropp, H. A. & Umar, A. Cancer prevention and screening: the next step in the era of precision medicine. NPJ Precis. Oncol. 3, 3 (2019).
Jeannot, E. et al. Circulating human papillomavirus DNA detected using droplet digital PCR in the serum of patients diagnosed with early stage human papillomavirus-associated invasive carcinoma. J. Pathol. Clin. Res. 2, 201–209 (2016).
Chan, K. C. A. et al. Analysis of plasma Epstein–Barr virus dna to screen for nasopharyngeal cancer. N. Engl. J. Med. 377, 513–522 (2017).
Abbosh, C., Birkbak, N. J. & Swanton, C. Early stage NSCLC—challenges to implementing ctDNA-based screening and MRD detection. Nat. Rev. Clin. Oncol. 15, 577–586 (2018).
Liu, M. C. et al. Breast cancer cell-free DNA (cfDNA) profiles reflect underlying tumor biology: the Circulating Cell-Free Genome Atlas (CCGA) study. J. Clin. Oncol. 36, 536–536 (2018).
Klein, E. A. et al. Development of a comprehensive cell-free DNA (cfDNA) assay for early detection of multiple tumor types: the Circulating Cell-free Genome Atlas (CCGA) study. J. Clin. Oncol. 36, 12021–12021 (2018).
Newman, A. M. et al. An ultrasensitive method for quantitating circulating tumor DNA with broad patient coverage. Nat. Med. 20, 548–554 (2014).
Newman, A. M. et al. Integrated digital error suppression for improved detection of circulating tumor DNA. Nat. Biotechnol. 34, 547–555 (2016).
Mouliere, F. et al. Enhanced detection of circulating tumor DNA by fragment size analysis. Sci. Transl. Med. 10, eaat4921 (2018).
Mansukhani, S. et al. Ultra-sensitive mutation detection and genome-wide DNA copy number reconstruction by error-corrected circulating tumor dna sequencing. Clin. Chem. 64, 1626–1635 (2018).
Yizhak, K. et al. RNA sequence analysis reveals macroscopic somatic clonal expansion across normal tissues. Science 364, eaaw0726 (2019).
Young, A. L., Challen, G. A., Birmann, B. M. & Druley, T. E. Clonal haematopoiesis harbouring AML-associated mutations is ubiquitous in healthy adults. Nat. Commun. 7, 12484 (2016).
Genovese, G. et al. Clonal hematopoiesis and blood-cancer risk inferred from blood DNA sequence. N. Engl. J. Med. 371, 2477–2487 (2014).
Mayrhofer, M. et al. Cell-free DNA profiling of metastatic prostate cancer reveals microsatellite instability, structural rearrangements and clonal hematopoiesis. Genome Med. 10, 85 (2018).
Fernandez-Cuesta, L. et al. Identification of circulating tumor DNA for the early detection of small-cell lung cancer. EBioMedicine 10, 117–123 (2016).
Cristiano, S. et al. Genome-wide cell-free DNA fragmentation in patients with cancer. Nature 570, 385–389 (2019).
Cohen, J. D. et al. Detection and localization of surgically resectable cancers with a multi-analyte blood test. Science 359, 926–930 (2018).
Pantel, K. & Alix-Panabieres, C. Liquid biopsy and minimal residual disease—latest advances and implications for cure. Nat. Rev. Clin. Oncol. 16, 409–424 (2019).
Riethdorf, S. et al. Prognostic impact of circulating tumor cells for breast cancer patients treated in the neoadjuvant “GeparQuattro” trial. Clin. Cancer Res. 23, 5384–5393 (2017).
Bidard, F. C. et al. Circulating tumor cells in breast cancer patients treated by neoadjuvant chemotherapy: a meta-analysis. J. Natl Cancer Inst. 110, 560–567 (2018).
Dasari, A., Grothey, A. & Kopetz, S. Circulating tumor DNA-defined minimal residual disease in solid tumors: opportunities to accelerate the development of adjuvant therapies. J. Clin. Oncol. 36, 3437–3440 (2018).
Lee, J. H. et al. Pre-operative ctDNA predicts survival in high-risk stage III cutaneous melanoma patients. Ann. Oncol. 30, 815–822 (2019).
Madic, J. et al. Circulating tumor DNA and circulating tumor cells in metastatic triple negative breast cancer patients. Int. J. Cancer 136, 2158–2165 (2015).
Rossi, G. et al. Cell-free DNA and circulating tumor cells: comprehensive liquid biopsy analysis in advanced breast cancer. Clin. Cancer Res. 24, 560–568 (2018).
Kasimir-Bauer, S. et al. Does primary neoadjuvant systemic therapy eradicate minimal residual disease? Analysis of disseminated and circulating tumor cells before and after therapy. Breast Cancer Res. 18, 20 (2016).
Strilic, B. & Offermanns, S. Intravascular survival and extravasation of tumor cells. Cancer Cell 32, 282–293 (2017).
Sparano, J. et al. Association of circulating tumor cells with late recurrence of estrogen receptor-positive breast cancer: a secondary analysis of a randomized clinical trial. JAMA Oncol. 4, 1700–1706 (2018).
Tie, J. et al. Circulating tumor DNA analysis detects minimal residual disease and predicts recurrence in patients with stage II colon cancer. Sci. Transl. Med. 8, 346ra392 (2016).
Gulbahce, N. et al. Quantitative whole genome sequencing of circulating tumor cells enables personalized combination therapy of metastatic cancer. Cancer Res. 77, 4530–4541 (2017).
Wikman, H. et al. Relevance of PTEN loss in brain metastasis formation in breast cancer patients. Breast Cancer Res. 14, R49 (2012).
Fluegen, G. et al. Phenotypic heterogeneity of disseminated tumour cells is preset by primary tumour hypoxic microenvironments. Nat. Cell Biol. 19, 120–132 (2017).
Aceto, N. et al. AR expression in breast cancer ctcs associates with bone metastases. Mol. Cancer Res. 16, 720–727 (2018).
Widschwendter, M. et al. The potential of circulating tumor DNA methylation analysis for the early detection and management of ovarian cancer. Genome Med. 9, 116 (2017).
Widschwendter, M. et al. Methylation patterns in serum DNA for early identification of disseminated breast cancer. Genome Med. 9, 115 (2017).
Moss, J. et al. Comprehensive human cell-type methylation atlas reveals origins of circulating cell-free DNA in health and disease. Nat. Commun. 9, 5068 (2018).
Hoadley, K. A. et al. Multiplatform analysis of 12 cancer types reveals molecular classification within and across tissues of origin. Cell 158, 929–944 (2014).
Warren, J. D. et al. Septin 9 methylated DNA is a sensitive and specific blood test for colorectal cancer. BMC Med. 9, 133 (2011).
Shen, S. Y. et al. Sensitive tumour detection and classification using plasma cell-free DNA methylomes. Nature 563, 579–583 (2018). This work shows the potential of plasma cfDNA methylation patterns to become a biomarker for the detection and classification of early-stage cancers.
Sun, K. et al. Orientation-aware plasma cell-free DNA fragmentation analysis in open chromatin regions informs tissue of origin. Genome Res. 29, 418–427 (2019).
Ulz, P. et al. Inferring expressed genes by whole-genome sequencing of plasma DNA. Nat. Genet. 48, 1273–1278 (2016). By performing whole-genome sequencing of plasma DNA, the authors could determine nucleosome occupancy and infer gene expression, which might help in the future to provide information about the location of origin of CTCs or ctDNA.
Paolillo, C. et al. Detection of activating estrogen receptor gene (ESR1) mutations in single circulating tumor cells. Clin. Cancer Res. 23, 6086–6093 (2017).
Sundaresan, T. K. et al. Detection of T790M, the acquired resistance EGFR mutation, by tumor biopsy versus noninvasive blood-based analyses. Clin. Cancer Res. 22, 1103–1110 (2016).
Misale, S. et al. Emergence of KRAS mutations and acquired resistance to anti-EGFR therapy in colorectal cancer. Nature 486, 532–536 (2012).
Oxnard, G. R. et al. Noninvasive detection of response and resistance in EGFR-mutant lung cancer using quantitative next-generation genotyping of cell-free plasma DNA. Clin. Cancer Res. 20, 1698–1705 (2014).
Murtaza, M. et al. Non-invasive analysis of acquired resistance to cancer therapy by sequencing of plasma DNA. Nature 497, 108–112 (2013).
Annala, M. et al. Circulating tumor DNA genomics correlate with resistance to abiraterone and enzalutamide in prostate cancer. Cancer Discov. 8, 444–457 (2018).
Bettegowda, C. et al. Detection of circulating tumor DNA in early- and late-stage human malignancies. Sci. Transl. Med. 6, 224ra224 (2014).
Parkinson, C. A. et al. Exploratory analysis of tp53 mutations in circulating tumour DNA as biomarkers of treatment response for patients with relapsed high-grade serous ovarian carcinoma: a retrospective study. PLOS Med. 13, e1002198 (2016).
Murtaza, M. et al. Multifocal clonal evolution characterized using circulating tumour DNA in a case of metastatic breast cancer. Nat. Commun. 6, 8760 (2015).
Russo, M. et al. Tumor heterogeneity and lesion-specific response to targeted therapy in colorectal cancer. Cancer Discov. 6, 147–153 (2016).
Wong, S. Q. et al. Circulating tumor DNA analysis and functional imaging provide complementary approaches for comprehensive disease monitoring in metastatic melanoma. JCO Precis. Oncol. https://doi.org/10.1200/PO.16.00009 (2017).
Merker, J. D. et al. Circulating tumor DNA analysis in patients with cancer: American Society of Clinical Oncology and College of American Pathologists joint review. J. Clin. Oncol. 36, 1631–1641 (2018).
Jahr, S. et al. DNA fragments in the blood plasma of cancer patients: quantitations and evidence for their origin from apoptotic and necrotic cells. Cancer Res. 61, 1659–1665 (2001).
Krug, A. K. et al. Improved EGFR mutation detection using combined exosomal RNA and circulating tumor DNA in NSCLC patient plasma. Ann. Oncol. 29, 700–706 (2018).
Markou, A. et al. Multiplex gene expression profiling of in vivo isolated circulating tumor cells in high-risk prostate cancer patients. Clin. Chem. 64, 297–306 (2018).
Dagogo-Jack, I. & Shaw, A. T. Tumour heterogeneity and resistance to cancer therapies. Nat. Rev. Clin. Oncol. 15, 81–94 (2018).
Dagogo-Jack, I. et al. Molecular analysis of plasma from patients with ROS1-positive NSCLC. J. Thorac. Oncol. 14, 816–824 (2019).
Pailler, E. et al. Circulating tumor cells with aberrant alk copy number predict progression-free survival during crizotinib treatment in alk-rearranged non-small cell lung cancer patients. Cancer Res. 77, 2222–2230 (2017).
Pailler, E. et al. High level of chromosomal instability in circulating tumor cells of ROS1-rearranged non-small-cell lung cancer. Ann. Oncol. 26, 1408–1415 (2015).
Dagogo-Jack, I. et al. Tracking the evolution of resistance to ALK tyrosine kinase inhibitors through longitudinal analysis of circulating tumor DNA. JCO Precis. Oncol. https://doi.org/10.1200/PO.17.00160 (2018).
Carter, L. et al. Molecular analysis of circulating tumor cells identifies distinct copy-number profiles in patients with chemosensitive and chemorefractory small-cell lung cancer. Nat. Med. 23, 114–119 (2017).
Hugo, W. et al. Non-genomic and immune evolution of melanoma acquiring MAPKi resistance. Cell 162, 1271–1285 (2015).
Scher, H. I. et al. Assessment of the validity of nuclear-localized androgen receptor splice variant 7 in circulating tumor cells as a predictive biomarker for castration-resistant prostate cancer. JAMA Oncol. 4, 1179–1186 (2018).
Jolly, M. K., Kulkarni, P., Weninger, K., Orban, J. & Levine, H. Phenotypic plasticity, bet-hedging, and androgen independence in prostate cancer: role of non-genetic heterogeneity. Front. Oncol. 8, 50 (2018).
Antonarakis, E. S. et al. Clinical significance of androgen receptor splice variant-7 mRNA detection in circulating tumor cells of men with metastatic castration-resistant prostate cancer treated with first- and second-line abiraterone and enzalutamide. J. Clin. Oncol. 35, 2149–2156 (2017). This study confirms the importance of CTC-based AR-V7 mRNA detection in predicting outcomes in patients with castration-resistant prostate cancer receiving first- and second-line anti-androgen therapy.
Miyamoto, D. T. et al. RNA-seq of single prostate CTCs implicates noncanonical Wnt signaling in antiandrogen resistance. Science 349, 1351–1356 (2015).
Tsao, S. C. et al. Characterising the phenotypic evolution of circulating tumour cells during treatment. Nat. Commun. 9, 1482 (2018).
Jordan, N. V. et al. HER2 expression identifies dynamic functional states within circulating breast cancer cells. Nature 537, 102–106 (2016). This study shows that CTCs from breast cancer that was originally ER + and HER – interconvert to a HER2 + phenotype under chemotherapy, which might have implications for switching therapy.
Chen, P. Y. et al. Adaptive and reversible resistance to Kras inhibition in pancreatic cancer cells. Cancer Res. 78, 985–1002 (2018).
Hata, A. N. et al. Tumor cells can follow distinct evolutionary paths to become resistant to epidermal growth factor receptor inhibition. Nat. Med. 22, 262–269 (2016).
Magbanua, M. J. M. et al. Expanded genomic profiling of circulating tumor cells in metastatic breast cancer patients to assess biomarker status and biology over time (CALGB 40502 and CALGB 40503, alliance). Clin. Cancer Res. 24, 1486–1499 (2018).
Drapkin, B. J. et al. Genomic and functional fidelity of small cell lung cancer patient-derived xenografts. Cancer Discov. 8, 600–615 (2018).
Yu, M. et al. Cancer therapy. Ex vivo culture of circulating breast tumor cells for individualized testing of drug susceptibility. Science 345, 216–220 (2014).
Franken, A. et al. Label-free enrichment and molecular characterization of viable circulating tumor cells from diagnostic leukapheresis products. Clin. Chem. 65, 549–558 (2019).
Gorges, T. M. et al. Enumeration and molecular characterization of tumor cells in lung cancer patients using a novel in vivo device for capturing circulating tumor cells. Clin. Cancer Res. 22, 2197–2206 (2016).
Bidard, F.-C. et al. Clinical utility of circulating tumor cell count as a tool to chose between first line hormone therapy and chemotherapy for ER+ HER2- metastatic breast cancer: results of the phase III STIC CTC trial. Cancer Res. 79 (4 Suppl.), abstr. GS3-07 (2018).
US National Library of Medicine. ClinicalTrials.gov, https://clinicaltrials.gov/ct2/show/NCT02035813 (2014).
US National Library of Medicine. ClinicalTrials.gov, https://clinicaltrials.gov/ct2/show/NCT03050866 (2017).
US National Library of Medicine. ClinicalTrials.gov, https://clinicaltrials.gov/ct2/show/NCT03844620 (2019).
US National Library of Medicine. ClinicalTrials.gov, https://clinicaltrials.gov/ct2/show/NCT03145961 (2017).
Wolchok, J. D. et al. Overall survival with combined nivolumab and ipilimumab in advanced melanoma. N. Engl. J. Med. 377, 1345–1356 (2017).
Reck, M. et al. Pembrolizumab versus chemotherapy for PD-L1-positive non-small-cell lung cancer. N. Engl. J. Med. 375, 1823–1833 (2016).
Ramskold, D. et al. Full-length mRNA-seq from single-cell levels of RNA and individual circulating tumor cells. Nat. Biotechnol. 30, 777–782 (2012).
Pantel, K. et al. Frequent down-regulation of major histocompatibility class I antigen expression on individual micrometastatic carcinoma cells. Cancer Res. 51, 4712–4715 (1991).
Mazel, M. et al. Frequent expression of PD-L1 on circulating breast cancer cells. Mol. Oncol. 9, 1773–1782 (2015).
Guibert, N. et al. PD-L1 expression in circulating tumor cells of advanced non-small cell lung cancer patients treated with nivolumab. Lung Cancer 120, 108–112 (2018).
Nicolazzo, C. et al. Monitoring PD-L1 positive circulating tumor cells in non-small cell lung cancer patients treated with the PD-1 inhibitor Nivolumab. Sci. Rep. 6, 31726 (2016).
Wang, Y. et al. PD-L1 expression in circulating tumor cells increases during radio(chemo)therapy and indicates poor prognosis in non-small cell lung cancer. Sci. Rep. 9, 566 (2019).
Strati, A. et al. Prognostic significance of PD-L1 expression on circulating tumor cells in patients with head and neck squamous cell carcinoma. Ann. Oncol. 28, 1923–1933 (2017).
Lee, J. H. et al. Circulating tumour DNA predicts response to anti-PD1 antibodies in metastatic melanoma. Ann. Oncol. 28, 1130–1136 (2017).
Cabel, L. et al. Clinical potential of circulating tumour DNA in patients receiving anticancer immunotherapy. Nat. Rev. Clin. Oncol. 15, 639–650 (2018).
Lee, J. H. et al. Association between circulating tumor DNA and pseudoprogression in patients with metastatic melanoma treated with anti-programmed cell death 1 antibodies. JAMA Oncol. 4, 717–721 (2018).
Gandara, D. R. et al. Blood-based tumor mutational burden as a predictor of clinical benefit in non-small-cell lung cancer patients treated with atezolizumab. Nat. Med. 24, 1441–1448 (2018).
Schumacher, T. N. & Schreiber, R. D. Neoantigens in cancer immunotherapy. Science 348, 69–74 (2015).
Del Re, M. et al. PD-L1 mRNA expression in plasma-derived exosomes is associated with response to anti-PD-1 antibodies in melanoma and NSCLC. Br. J. Cancer 118, 820–824 (2018).
Ricklefs, F. L. et al. Immune evasion mediated by PD-L1 on glioblastoma-derived extracellular vesicles. Sci. Adv. 4, eaar2766 (2018).
Chen, G. et al. Exosomal PD-L1 contributes to immunosuppression and is associated with anti-PD-1 response. Nature 560, 382–386 (2018). This study unravels a mechanism by which tumour cells systematically suppress the immune system and suggests exosomal PDL1 as a biomarker of clinical response to immune checkpoint inhibition therapy.
Gabrilovich, D. I. Myeloid-derived suppressor cells. Cancer Immunol. Res. 5, 3–8 (2017).
Ferrucci, P. F. et al. Baseline neutrophils and derived neutrophil-to-lymphocyte ratio: prognostic relevance in metastatic melanoma patients receiving ipilimumab. Ann. Oncol. 27, 732–738 (2016).
Huber, V. et al. Tumor-derived microRNAs induce myeloid suppressor cells and predict immunotherapy resistance in melanoma. J. Clin. Invest. 128, 5505–5516 (2018).
Boeri, M. et al. Circulating miRNAs and PD-L1 tumor expression are associated with survival in advanced NSCLC patients treated with immunotherapy: a prospective study. Clin. Cancer Res. 25, 2166–2173 (2019).
Merino, D. et al. Barcoding reveals complex clonal behavior in patient-derived xenografts of metastatic triple negative breast cancer. Nat. Commun. 10, 766 (2019).
Stoeckius, M. et al. Simultaneous epitope and transcriptome measurement in single cells. Nat. Methods 14, 865–868 (2017).
Clark, S. J. et al. scNMT-seq enables joint profiling of chromatin accessibility DNA methylation and transcription in single cells. Nat. Commun. 9, 781 (2018).
Théry, C. et al. Minimal information for studies of extracellular vesicles 2018 (MISEV2018): a position statement of the International Society for Extracellular Vesicles and update of the MISEV2014 guidelines. J. Extracell. Vesicles 7, 1535750 (2018).
Best, M. G. et al. Swarm intelligence-enhanced detection of non-small-cell lung cancer using tumor-educated platelets. Cancer Cell 32, 238–252.e239 (2017). This study demonstrates accurate tumour-educated platelet-based detection of early- and late-stage NSCLC with the use of RNA sequencing and an optimized classification algorithm.
Tkach, M. & Thery, C. Communication by extracellular vesicles: where we are and where we need to go. Cell 164, 1226–1232 (2016).
Ko, J. et al. Machine learning to detect signatures of disease in liquid biopsies—a user’s guide. Lab Chip 18, 395–405 (2018).
Elias, K. M. et al. Diagnostic potential for a serum miRNA neural network for detection of ovarian cancer. eLife 6, e28932 (2017).
Nitta, N. et al. Intelligent image-activated cell sorting. Cell 175, 266–276.e213 (2018).
Brasko, C. et al. Intelligent image-based in situ single-cell isolation. Nat. Commun. 9, 226 (2018).
Chen, C. L. et al. Deep learning in label-free cell classification. Sci. Rep. 6, 21471 (2016).
Snyder, M. W., Kircher, M., Hill, A. J., Daza, R. M. & Shendure, J. Cell-free DNA comprises an in vivo nucleosome footprint that informs its tissues-of-origin. Cell 164, 57–68 (2016).
Yuan, T. et al. Plasma extracellular RNA profiles in healthy and cancer patients. Sci. Rep. 6, 19413 (2016).
Tkach, M. et al. Qualitative differences in T-cell activation by dendritic cell-derived extracellular vesicle subtypes. EMBO J. 36, 3012–3028 (2017).
Plantureux, L. et al. Impacts of cancer on platelet production, activation and education and mechanisms of cancer-associated thrombosis. Cancers 10, E441 (2018).
In ‘t Veld, S. & Wurdinger, T. Tumor-educated platelets. Blood 133, 2359–2364 (2019).
Poudineh, M., Sargent, E. H., Pantel, K. & Kelley, S. O. Profiling circulating tumour cells and other biomarkers of invasive cancers. Nat. Biomed. Eng. 2, 72–84 (2018).
Park, E. S. et al. Isolation and genome sequencing of individual circulating tumor cells using hydrogel encapsulation and laser capture microdissection. Lab Chip 18, 1736–1749 (2018).
Babayan, A. et al. Comparative study of whole genome amplification and next generation sequencing performance of single cancer cells. Oncotarget 8, 56066–56080 (2017).
Rihawi, K. et al. MYC amplification as a potential mechanism of primary resistance to crizotinib in ALK-rearranged non-small cell lung cancer: a brief report. Transl. Oncol. 12, 116–121 (2019).
Berger, L.-A. et al. Identification of a high-level MET amplification in CTCs and cfTNA of an ALK-positive NSCLC patient developing evasive resistance to crizotinib. J. Thorac. Oncol. 13, e243–e246 (2018).
Valihrach, L., Androvic, P. & Kubista, M. Platforms for single-cell collection and analysis. Int. J. Mol. Sci. 19, E807 (2018).
Castro-Giner, F., Scheidmann, M. C. & Aceto, N. Beyond enumeration: functional and computational analysis of circulating tumor cells to investigate cancer metastasis. Front. Med. 5, 34 (2018).
Heather, J. M. & Chain, B. The sequence of sequencers: the history of sequencing DNA. Genomics 107, 1–8 (2016).
Fehm, T. et al. Cytogenetic evidence that circulating epithelial cells in patients with carcinoma are malignant. Clin. Cancer Res. 8, 2073–2084 (2002).
Lee, J. H. et al. Fluorescent in situ sequencing (FISSEQ) of RNA for gene expression profiling in intact cells and tissues. Nat. Protoc. 10, 442–458 (2015).
Mazor, T., Pankov, A., Song, J. S. & Costello, J. F. Intratumoral heterogeneity of the epigenome. Cancer Cell 29, 440–451 (2016).
Pixberg, C. F. et al. Analysis of DNA methylation in single circulating tumor cells. Oncogene 36, 3223–3231 (2017).
Kelsey, G., Stegle, O. & Reik, W. Single-cell epigenomics: recording the past and predicting the future. Science 358, 69–75 (2017).
Baslan, T. & Hicks, J. Unravelling biology and shifting paradigms in cancer with single-cell sequencing. Nat. Rev. Cancer 17, 557–569 (2017).
Gawad, C., Koh, W. & Quake, S. R. Single-cell genome sequencing: current state of the science. Nat. Rev. Genet. 17, 175–188 (2016).
Kolodziejczyk, A. A., Kim, J. K., Svensson, V., Marioni, J. C. & Teichmann, S. A. The technology and biology of single-cell RNA sequencing. Mol. Cell 58, 610–620 (2015).
Sinkala, E. et al. Profiling protein expression in circulating tumour cells using microfluidic western blotting. Nat. Commun. 8, 14622 (2017).
Spitzer, M. H. & Nolan, G. P. Mass cytometry: single cells, many features. Cell 165, 780–791 (2016).
Gerdtsson, E. et al. Multiplex protein detection on circulating tumor cells from liquid biopsies using imaging mass cytometry. Converg. Sci. Phys. Oncol. 4, 015002 (2018).
Zenobi, R. Single-cell metabolomics: analytical and biological perspectives. Science 342, 1243259 (2013).
Del Ben, F. et al. A method for detecting circulating tumor cells based on the measurement of single-cell metabolism in droplet-based microfluidics. Angew. Chem. Int. Ed. Engl. 55, 8581–8584 (2016).
Nieto, M. A., Huang, R. Y., Jackson, R. A. & Thiery, J. P. EMT: 2016. Cell 166, 21–45 (2016).
Yokobori, T. et al. Plastin3 is a novel marker for circulating tumor cells undergoing the epithelial–mesenchymal transition and is associated with colorectal cancer prognosis. Cancer Res. 73, 2059–2069 (2013).
Agerbaek, M. O. et al. The VAR2CSA malaria protein efficiently retrieves circulating tumor cells in an EpCAM-independent manner. Nat. Commun. 9, 3279 (2018).
Riethdorf, S., O’Flaherty, L., Hille, C. & Pantel, K. Clinical applications of the CellSearch platform in cancer patients. Adv. Drug Deliv. Rev. 125, 102–121 (2018).
Agnoletto, C. et al. Heterogeneity in circulating tumor cells: the relevance of the stem-cell subset. Cancers 11, E483 (2019).
Xu, L. et al. The novel association of circulating tumor cells and circulating megakaryocytes with prostate cancer prognosis. Clin. Cancer Res. 23, 5112–5122 (2017). This study demonstrates that the majority of circulating cytokeratin-negative, vimentin-positive and CD45-negative cells, which could be mesenchymal CTCs or endothelial cells, have tumour-specific genomic aberrations in prostate cancer.
Morrow, C. J. et al. Tumourigenic non-small-cell lung cancer mesenchymal circulating tumour cells: a clinical case study. Ann. Oncol. 27, 1155–1160 (2016).
Comaills, V. et al. Genomic instability is induced by persistent proliferation of cells undergoing epithelial-to-mesenchymal transition. Cell Rep. 17, 2632–2647 (2016).
Sun, Y. F. et al. Circulating tumor cells from different vascular sites exhibit spatial heterogeneity in epithelial and mesenchymal composition and distinct clinical significance in hepatocellular carcinoma. Clin. Cancer Res. 24, 547–559 (2018).
Labelle, M., Begum, S. & Hynes, R. O. Direct signaling between platelets and cancer cells induces an epithelial–mesenchymal-like transition and promotes metastasis. Cancer Cell 20, 576–590 (2011).
Tam, W. L. & Weinberg, R. A. The epigenetics of epithelial–mesenchymal plasticity in cancer. Nat. Med. 19, 1438–1449 (2013).
Galanzha, E. I. et al. In vivo liquid biopsy using Cytophone platform for photoacoustic detection of circulating tumor cells in patients with melanoma. Sci. Transl. Med. 11, eaat5857 (2019).
Vermesh, O. et al. An intravascular magnetic wire for the high-throughput retrieval of circulating tumour cells in vivo. Nat. Biomed. Eng. 2, 696–705 (2018).
Kim, T. H. et al. A temporary indwelling intravascular aphaeretic system for in vivo enrichment of circulating tumor cells. Nat. Commun. 10, 1478 (2019).
Hamza, B. et al. Optofluidic real-time cell sorter for longitudinal CTC studies in mouse models of cancer. Proc. Natl Acad. Sci. USA 116, 2232–2236 (2019).
Weiss, L. et al. Haematogenous metastatic patterns in colonic carcinoma: an analysis of 1541 necropsies. J. Pathol. 150, 195–203 (1986).
Weiss, L. et al. Metastatic patterns of renal carcinoma: an analysis of 687 necropsies. J. Cancer Res. Clin. Oncol. 114, 605–612 (1988).
Maestro, L. M. et al. Circulating tumor cells in solid tumor in metastatic and localized stages. Anticancer Res. 29, 4839–4843 (2009).
Lucci, A. et al. Circulating tumour cells in non-metastatic breast cancer: a prospective study. Lancet Oncol. 13, 688–695 (2012).
Cristofanilli, M. et al. Circulating tumor cells, disease progression, and survival in metastatic breast cancer. N. Engl. J. Med. 351, 781–791 (2004).
Thalgott, M. et al. Detection of circulating tumor cells in different stages of prostate cancer. J. Cancer Res. Clin. Oncol. 139, 755–763 (2013).
Andree, K. C. et al. Toward a real liquid biopsy in metastatic breast and prostate cancer: diagnostic leukapheresis increases CTC yields in a European prospective multicenter study (CTCTrap). Int. J. Cancer 143, 2584–2591 (2018).
Stoecklein, N. H., Fischer, J. C., Niederacher, D. & Terstappen, L. W. Challenges for CTC-based liquid biopsies: low CTC frequency and diagnostic leukapheresis as a potential solution. Expert Rev. Mol. Diagn. 16, 147–164 (2016).
Acknowledgements
K.P. received research funding from the Deutsche Forschungsgemeinschaft (DFG) SPP2084 µBone, Deutsche Krebshilfe 'Translationale Onkologie' n° 70112507, EU TRANSCAN-199 Grant PROLIPSY, ERC PoC Grant CTCapture_2.0 n°7544533 H2020 and the EU/IMI consortium CANCER-ID under grant agreement n° 115749, European Union’s Seventh Framework Programme (FP7/2007e2013). L.K. was financially supported by ITMO Cancer 'Plan Cancer 2014-2019' and received funding from Fondation de France.
Author information
Authors and Affiliations
Contributions
Both authors contributed equally to all aspects of the article.
Corresponding author
Ethics declarations
Competing interests
K.P. has ongoing patent applications related to circulating tumour cells, and has received honoraria from Agena, Novartis, Roche and Sanofi and research funding from European Federation of Pharmaceutical Industries and Associations (EFPIA) partners (Angle, Menarini and Servier) of the CANCER-ID programme of the European Union–EFPIA Innovative Medicines Initiative. L.K. declares no competing interests.
Additional information
Peer review information
Nature Reviews Cancer thanks N. Aceto, S. Jeffrey and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Related links
Blood Profiling Atlas in Cancer (BloodPAC) consortium: https://www.bloodpac.org/
EU Innovative Medicines Initiative (IMI) consortium CANCER-ID: https://www.cancer-id.eu/
European Liquid Biopsy Society (ELBS): http://www.elbs.eu
Glossary
- Disseminated tumour cells
-
(DTCs). A fraction of circulating tumour cells that have extravasated through the endothelium of the blood vessels at distant sites.
- Micrometastases
-
A small collection of tumour cells that have metastasized from the primary tumour in different organs and that are not clinically detectable.
- Enriched fractions
-
Resulting pool of circulating tumour cells in a background of leukocytes after the capture method.
- Leptomeningeal metastasis
-
Corresponds to the spreading of tumour cells to leptomeninges (a thin layer of tissues that protect and cover the brain and the spinal cord).
- Cancer-associated fibroblasts
-
Stromal cells that populate the tumour microenvironment and are involved in cancer progression
- CTC-derived xenografts
-
(CDXs). A model generated from tumorigenic circulating tumour cells in immunocompromised mice.
- Parallel progression
-
A model of progression implying that a metastatic clone leaves the primary tumour early and continues to evolve in parallel to the primary tumour.
- Linear progression
-
A model of progression implying that cancer cells accumulate multiple successive rounds of mutation and selection for invasiveness within the primary tumour before metastasizing.
- Convergent evolution
-
The recurrent tendency of a biological system to independently evolve similar traits arriving at the same solution.
- Bisulfite conversion
-
A technique used to study DNA methylation. Following bisulphite treatment, cytosine is converted into uracil but methylated cytosine is left intact.
- Phenotypic switching
-
The ability of cells in a population to switch states (phenotypes) in response to environmental conditions despite having identical genetic contents.
- Optical loss
-
The attenuation of multiplexed wavelength components passing through the cells.
Rights and permissions
About this article
Cite this article
Keller, L., Pantel, K. Unravelling tumour heterogeneity by single-cell profiling of circulating tumour cells. Nat Rev Cancer 19, 553–567 (2019). https://doi.org/10.1038/s41568-019-0180-2
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41568-019-0180-2
This article is cited by
-
Histone 3 lysine 9 acetylation-specific reprogramming regulates esophageal squamous cell carcinoma progression and metastasis
Cancer Gene Therapy (2024)
-
Trastuzumab and first-line taxane chemotherapy in metastatic breast cancer patients with a HER2-negative tumor and HER2-positive circulating tumor cells: a phase II trial
Breast Cancer Research and Treatment (2024)
-
Clinical applications of circulating tumor cells in patients with solid tumors
Clinical & Experimental Metastasis (2024)
-
Research progress on the multi-omics and survival status of circulating tumor cells
Clinical and Experimental Medicine (2024)
-
Liquid biopsy techniques and pancreatic cancer: diagnosis, monitoring, and evaluation
Molecular Cancer (2023)