Abstract
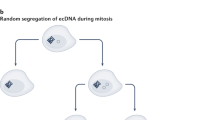
Recent reports have demonstrated that oncogene amplification on extrachromosomal DNA (ecDNA) is a frequent event in cancer, providing new momentum to explore a phenomenon first discovered several decades ago. The direct consequence of ecDNA gains in these cases is an increase in DNA copy number of the oncogenes residing on the extrachromosomal element. A secondary effect, perhaps even more important, is that the unequal segregation of ecDNA from a parental tumour cell to offspring cells rapidly increases tumour heterogeneity, thus providing the tumour with an additional array of responses to microenvironment-induced and therapy-induced stress factors and perhaps providing an evolutionary advantage. This Perspectives article discusses the current knowledge and potential implications of oncogene amplification on ecDNA in cancer.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Beroukhim, R. et al. The landscape of somatic copy-number alteration across human cancers. Nature 463, 899–905 (2010).
Weischenfeldt, J. et al. Pan-cancer analysis of somatic copy-number alterations implicates IRS4 and IGF2 in enhancer hijacking. Nat. Genet. 49, 65–74 (2017).
Menghi, F. et al. The tandem duplicator phenotype is a prevalent genome-wide cancer configuration driven by distinct gene mutations. Cancer Cell 34, 197–210 (2018).
Gisselsson, D. et al. Generation of trisomies in cancer cells by multipolar mitosis and incomplete cytokinesis. Proc. Natl Acad. Sci. USA 107, 20489–20493 (2010).
Korbel, J. O. & Campbell, P. J. Criteria for inference of chromothripsis in cancer genomes. Cell 152, 1226–1236 (2013).
Solomon, E., Borrow, J. & Goddard, A. D. Chromosome aberrations and cancer. Science 254, 1153–1160 (1991).
deCarvalho, A. C. et al. Discordant inheritance of chromosomal and extrachromosomal DNA elements contributes to dynamic disease evolution in glioblastoma. Nat. Genet. 50, 708–717 (2018).
Turner, K. M. et al. Extrachromosomal oncogene amplification drives tumour evolution and genetic heterogeneity. Nature 543, 122–125 (2017).
Garsed, D. W. et al. The architecture and evolution of cancer neochromosomes. Cancer Cell 26, 653–667 (2014).
Macchia, G. et al. The hidden genomic and transcriptomic plasticity of giant marker chromosomes in cancer. Genetics 208, 951–961 (2018).
Liehr, T., Claussen, U. & Starke, H. Small supernumerary marker chromosomes (sSMC) in humans. Cytogenet. Genome Res. 107, 55–67 (2004).
Paulsen, T., Kumar, P., Koseoglu, M. M. & Dutta, A. Discoveries of extrachromosomal circles of DNA in normal and tumor cells. Trends Genet. 34, 270–278 (2018).
Moller, H. D., Parsons, L., Jorgensen, T. S., Botstein, D. & Regenberg, B. Extrachromosomal circular DNA is common in yeast. Proc. Natl Acad. Sci. USA 112, E3114–E3122 (2015).
Stanfield, S. W. & Lengyel, J. A. Small circular DNA of Drosophila melanogaster: chromosomal homology and kinetic complexity. Proc. Natl Acad. Sci. USA 76, 6142–6146 (1979).
Shoura, M. J. et al. Intricate and cell type-specific populations of endogenous circular DNA (eccDNA) in Caenorhabditis elegans and Homo sapiens. G3 7, 3295–3303 (2017).
Shibata, Y. et al. Extrachromosomal microDNAs and chromosomal microdeletions in normal tissues. Science 336, 82–86 (2012).
Hotta, Y. & Bassel, A. Molecular size and circularity of DNA in cells of mammals and higher plants. Proc. Natl Acad. Sci. USA 53, 356–362 (1965).
Moller, H. D. et al. Circular DNA elements of chromosomal origin are common in healthy human somatic tissue. Nat. Commun. 9, 1069 (2018).
Henson, J. D. et al. DNA C-circles are specific and quantifiable markers of alternative-lengthening-of-telomeres activity. Nat. Biotechnol. 27, 1181–1185 (2009).
Cohen, S., Regev, A. & Lavi, S. Small polydispersed circular DNA (spcDNA) in human cells: association with genomic instability. Oncogene 14, 977–985 (1997).
Cox, D., Yuncken, C. & Spriggs, A. I. Minute chromatin bodies in malignant tumours of childhood. Lancet 1, 55–58 (1965).
Fan, Y. et al. Frequency of double minute chromosomes and combined cytogenetic abnormalities and their characteristics. J. Appl. Genet. 52, 53–59 (2011).
Stark, G. R., Debatisse, M., Giulotto, E. & Wahl, G. M. Recent progress in understanding mechanisms of mammalian DNA amplification. Cell 57, 901–908 (1989).
Schimke, R. T. Gene amplification in cultured animal cells. Cell 37, 705–713 (1984).
Alt, F. W., Kellems, R. E., Bertino, J. R. & Schimke, R. T. Selective multiplication of dihydrofolate reductase genes in methotrexate-resistant variants of cultured murine cells. J. Biol. Chem. 253, 1357–1370 (1978).
Haber, D. A., Beverley, S. M., Kiely, M. L. & Schimke, R. T. Properties of an altered dihydrofolate reductase encoded by amplified genes in cultured mouse fibroblasts. J. Biol. Chem. 256, 9501–9510 (1981).
Haber, D. A. & Schimke, R. T. Unstable amplification of an altered dihydrofolate reductase gene associated with double-minute chromosomes. Cell 26, 355–362 (1981).
Beverley, S. M., Coderre, J. A., Santi, D. V. & Schimke, R. T. Unstable DNA amplifications in methotrexate-resistant Leishmania consist of extrachromosomal circles which relocalize during stabilization. Cell 38, 431–439 (1984).
Kohl, N. E. et al. Transposition and amplification of oncogene-related sequences in human neuroblastomas. Cell 35, 359–367 (1983).
Alitalo, K., Schwab, M., Lin, C. C., Varmus, H. E. & Bishop, J. M. Homogeneously staining chromosomal regions contain amplified copies of an abundantly expressed cellular oncogene (c-myc) in malignant neuroendocrine cells from a human colon carcinoma. Proc. Natl Acad. Sci. USA 80, 1707–1711 (1983).
Von Hoff, D. D. et al. Elimination of extrachromosomally amplified MYC genes from human tumor cells reduces their tumorigenicity. Proc. Natl Acad. Sci. USA 89, 8165–8169 (1992).
Von Hoff, D. D. et al. Hydroxyurea accelerates loss of extrachromosomally amplified genes from tumor cells. Cancer Res. 51, 6273–6279 (1991).
Carroll, S. M. et al. Double minute chromosomes can be produced from precursors derived from a chromosomal deletion. Mol. Cell. Biol. 8, 1525–1533 (1988).
Ruiz, J. C., Choi, K. H., von Hoff, D. D., Roninson, I. B. & Wahl, G. M. Autonomously replicating episomes contain mdr1 genes in a multidrug-resistant human cell line. Mol. Cell. Biol. 9, 109–115 (1989).
Storlazzi, C. T. et al. MYC-containing double minutes in hematologic malignancies: evidence in favor of the episome model and exclusion of MYC as the target gene. Hum. Mol. Genet. 15, 933–942 (2006).
Storlazzi, C. T. et al. Gene amplification as double minutes or homogeneously staining regions in solid tumors: origin and structure. Genome Res. 20, 1198–1206 (2010).
Garraway, L. A. & Lander, E. S. Lessons from the cancer genome. Cell 153, 17–37 (2013).
Vogelstein, B. et al. Cancer genome landscapes. Science 339, 1546–1558 (2013).
Mitelman, F., Johansson, B. & Mertens, F. The impact of translocations and gene fusions on cancer causation. Nat. Rev. Cancer 7, 233–245 (2007).
Nathanson, D. A. et al. Targeted therapy resistance mediated by dynamic regulation of extrachromosomal mutant EGFR DNA. Science 343, 72–76 (2014).
Snuderl, M. et al. Mosaic amplification of multiple receptor tyrosine kinase genes in glioblastoma. Cancer Cell 20, 810–817 (2011).
Szerlip, N. J. et al. Intratumoral heterogeneity of receptor tyrosine kinases EGFR and PDGFRA amplification in glioblastoma defines subpopulations with distinct growth factor response. Proc. Natl Acad. Sci. USA 109, 3041–3046 (2012).
Vogt, N. et al. Molecular structure of double-minute chromosomes bearing amplified copies of the epidermal growth factor receptor gene in gliomas. Proc. Natl Acad. Sci. USA 101, 11368–11373 (2004).
Bigner, S. H. et al. Relationship between gene amplification and chromosomal deviations in malignant human gliomas. Cancer Genet. Cytogenet. 29, 165–170 (1987).
Deshpande, V. et al. Exploring the landscape of focal amplifications in cancer using AmpliconArchitect. Nat. Commun. 10, 392 (2019).
L’Abbate, A. et al. Genomic organization and evolution of double minutes/homogeneously staining regions with MYC amplification in human cancer. Nucleic Acids Res. 42, 9131–9145 (2014).
Ly, P. & Cleveland, D. W. Rebuilding chromosomes after catastrophe: emerging mechanisms of chromothripsis. Trends Cell Biol. 27, 917–930 (2017).
Zheng, S. et al. A survey of intragenic breakpoints in glioblastoma identifies a distinct subset associated with poor survival. Genes Dev. 27, 1462–1472 (2013).
Malhotra, A. et al. Breakpoint profiling of 64 cancer genomes reveals numerous complex rearrangements spawned by homology-independent mechanisms. Genome Res. 23, 762–776 (2013).
Xu, K. et al. Structure and evolution of double minutes in diagnosis and relapse brain tumors. Acta Neuropathol. 137, 123–137 (2018).
Nikolaev, S. et al. Extrachromosomal driver mutations in glioblastoma and low-grade glioma. Nat. Commun. 5, 5690 (2014).
Yost, S. E. et al. High-resolution mutational profiling suggests the genetic validity of glioblastoma patient-derived pre-clinical models. PLOS ONE 8, e56185 (2013).
Xue, Y. et al. An approach to suppress the evolution of resistance in BRAF(V600E)-mutant cancer. Nat. Med. 23, 929–937 (2017).
Gatenby, R. A. & Gillies, R. J. A microenvironmental model of carcinogenesis. Nat. Rev. Cancer 8, 56–61 (2008).
Zhang, A. W. et al. Interfaces of malignant and immunologic clonal dynamics in ovarian cancer. Cell 173, 1755–1769 (2018).
Grasso, C. S. et al. Genetic mechanisms of immune evasion in colorectal cancer. Cancer Discov. 8, 730–749 (2018).
McGranahan, N. et al. Allele-specific HLA loss and immune escape in lung cancer evolution. Cell 171, 1259–1271 (2017).
Whyte, W. A. et al. Master transcription factors and mediator establish super-enhancers at key cell identity genes. Cell 153, 307–319 (2013).
Dixon, J. R. et al. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature 485, 376–380 (2012).
Shachar, S., Voss, T. C., Pegoraro, G., Sciascia, N. & Misteli, T. Identification of gene positioning factors using high-throughput imaging mapping. Cell 162, 911–923 (2015).
Hnisz, D. et al. Activation of proto-oncogenes by disruption of chromosome neighborhoods. Science 351, 1454–1458 (2016).
Flavahan, W. A. et al. Insulator dysfunction and oncogene activation in IDH mutant gliomas. Nature 529, 110–114 (2016).
Corces, M. R. et al. The chromatin accessibility landscape of primary human cancers. Science 362, eaav1898 (2018).
Moller, H. D. et al. CRISPR-C: circularization of genes and chromosome by CRISPR in human cells. Nucleic Acids Res. 46, e131 (2018).
Grohmann, E., Muth, G. & Espinosa, M. Conjugative plasmid transfer in gram-positive bacteria. Microbiol. Mol. Biol. Rev. 67, 277–301 (2003).
Figurski, D. H. & Helinski, D. R. Replication of an origin-containing derivative of plasmid RK2 dependent on a plasmid function provided in trans. Proc. Natl Acad. Sci. USA 76, 1648–1652 (1979).
Bennett, P. M. Plasmid encoded antibiotic resistance: acquisition and transfer of antibiotic resistance genes in bacteria. Br. J. Pharmacol. 153 (Suppl. 1), 347–357 (2008).
Acknowledgements
The authors thank C. Beck (Jackson Laboratory for Genomic Medicine), S. Wu and K. M. Turner (Mischel laboratory) for feedback on the manuscript content and S. Cassidy (Jackson Laboratory for Genomic Medicine) for support in manuscript writing. R.G.W.V. is supported by grants from the US National Institutes of Health (NIH) (R01 CA190121), Cancer Center Support Grant P30CA034196 and grants from the Musella Foundation and the B*CURED Foundation. V.B. is supported in part by grants from the NIH (GM114362 and HG004962) and the US National Science Foundation (NSF) (DBI-1458557). P.S.M. is supported in part by grants from the NIH (NS73831), the Defeat GBM Program of the US National Brain Tumor Society, the Ben and Catherine Ivy Foundation, an award from the Sharpe–National Brain Tumor Society Research Program and a Compute for the Cure Award from the Nvidia Foundation.
Reviewer information
Nature Reviews Cancer thanks A. Dutta, W. Hahn and other anonymous reviewer(s) for their contribution to the peer review of this work.
Author information
Authors and Affiliations
Contributions
The authors contributed equally to all aspects of the article.
Corresponding authors
Ethics declarations
Competing interests
R.G.W.V., V.B. and P.S.M. are co-founders of and have equity interest in Pretzel Therapeutics (PT). P.S.M. serves as a consultant to PT. V.B. is a co-founder of, has equity interest in and receives income from Digital Proteomics (DP). The terms of this arrangement have been reviewed and approved by the University of California, San Diego, in accordance with its conflict of interest policies. PT and DP were not involved in the research presented here.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Glossary
- Alternative lengthening of telomeres
-
One or more mechanisms that are frequently observed in tumours lacking telomerase activity and manifested by long but highly variable telomeres.
- Breakage–fusion–bridge cycles
-
A mechanism of chromosomal instability initiated by a telomeric loss and multiple cycles of anaphase bridge formation followed by unequal breakage.
- Chromothripsis
-
A phenomenon marked by shattering of a chromosome and re-ligation of some of the fragments, causing massive rearrangement in a single catastrophic event.
- High-throughput short-read DNA sequencing
-
The term given to a variety of sequencing technologies that generate short (100–300 bp) reads in an unbiased and massively parallel manner, allowing for inexpensive and redundant sampling of a genome.
- Homogeneously staining regions
-
(HSRs). Regions of a chromosome that have duplicated many times and show up as large regions that are homogeneously stained when painted with a fluorescence in situ hybridization (FISH) probe unique to the region. While tandem duplications are usually implicated in HSR formation, replication of ecDNA and their reintegration into the genome may also cause HSRs.
- MicroDNAs
-
A form of short, extrachromosomal, circular DNA elements that are up to 400 bp long, non-repetitive and putatively formed owing to excision and replication of short DNA.
- Optical map technologies
-
Genomic technologies that construct ordered maps of restriction site locations in large genomic fragments (150–400 kb). The maps serve as a unique fingerprint for the fragment and are useful in validating large structural variations, including insertions.
- Supernumerary marker chromosomes
-
A phenomenon that occurs when cells have an additional and structurally abnormal copy of an autosomal chromosome. They are infrequently found in individuals.
- Tandem duplication
-
Repeated segments of DNA inserted in the genome that disrupt expression of important tumour suppressor genes or amplify tumour promoter genes.
- Telomeric circles
-
Structures based on circularization of telomeric tandem repeats that allow for rolling circle amplification and synthesis of longer repeat elements that help with lengthening of telomeres and stabilizing the chromosome.
- Topologically associating domain
-
A region of the genome that is characterized by extensive interactions within owing to the spatial organization of the genome and reduced interactions with regions outside.
Rights and permissions
About this article
Cite this article
Verhaak, R.G.W., Bafna, V. & Mischel, P.S. Extrachromosomal oncogene amplification in tumour pathogenesis and evolution. Nat Rev Cancer 19, 283–288 (2019). https://doi.org/10.1038/s41568-019-0128-6
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41568-019-0128-6
This article is cited by
-
Aneuploidy and complex genomic rearrangements in cancer evolution
Nature Cancer (2024)
-
Thinking outside the chromosome: epigenetic mechanisms in non-canonical chromatin species
Nature Structural & Molecular Biology (2024)
-
Imaging extrachromosomal DNA (ecDNA) in cancer
Histochemistry and Cell Biology (2024)
-
Dynamic genomic changes in methotrexate-resistant human cancer cell lines beyond DHFR amplification suggest potential new targets for preventing drug resistance
British Journal of Cancer (2024)
-
Small extrachromosomal circular DNA harboring targeted tumor suppressor gene mutations supports intratumor heterogeneity in mouse liver cancer induced by multiplexed CRISPR/Cas9
Genome Medicine (2023)