Abstract
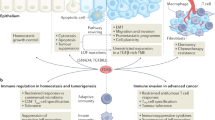
The tightly orchestrated temporal and spatial control of signal transducer and activator of transcription 3 (STAT3) activity in epithelial, immune and stromal cells is critical for wound healing and tissue repair. Excessive STAT3 activation within cancer cells and cells of the tumour microenvironment can be viewed as a neoplastic mimic of an inflammation-driven repair response that collectively promotes tumour progression. In addition to the canonical transcriptional pathways by which STAT3 promotes stem cell-like characteristics, survival, proliferation, metastatic potential and immune evasion, cytoplasmic STAT3 activity fuels tumour growth by metabolic and other non-transcriptional mechanisms. Here, we review the tumour-modulating activities of STAT3 in light of its role as a signalling node integrating inflammatory responses during wound healing. Accordingly, many of the cytokines that contribute to the para-inflammatory state of most solid malignancies converge on and underpin dysregulated STAT3 activity. Targeting of these cytokines, their cognate receptors and associated signalling cascades in clinical trials is beginning to demonstrate therapeutic efficacy, given that interference with STAT3 activity is likely to simultaneously curb the growth of cancer cells and augment antitumour immunity.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
O’Shea, J. J. et al. The JAK-STAT pathway: impact on human disease and therapeutic intervention. Annu. Rev. Med. 66, 311–328 (2015).
Heinrich, P. C. et al. Principles of interleukin (IL)-6-type cytokine signalling and its regulation. Biochem. J. 374, 1–20 (2003).
Groner, B. & von Manstein, V. Jak Stat signaling and cancer: opportunities, benefits and side effects of targeted inhibition. Mol. Cell. Endocrinol. 451, 1–14 (2017).
Yu, H., Lee, H., Herrmann, A., Buettner, R. & Jove, R. Revisiting STAT3 signalling in cancer: new and unexpected biological functions. Nat. Rev. Cancer 14, 736–746 (2014).
Johnson, D. E., O’Keefe, R. A. & Grandis, J. R. Targeting the IL-6/JAK/STAT3 signalling axis in cancer. Nat. Rev. Clin. Oncol. 15, 234–248 (2018).
Hillmer, E. J., Zhang, H., Li, H. S. & Watowich, S. S. STAT3 signaling in immunity. Cytokine Growth Factor Rev. 31, 1–15 (2016).
Dvorak, H. F. Tumors: wounds that do not heal. Similarities between tumor stroma generation and wound healing. N. Engl. J. Med. 315, 1650–1659 (1986).
Hanahan, D. & Weinberg, R. A. Hallmarks of cancer: the next generation. Cell 144, 646–674 (2011).
Sano, S. et al. Keratinocyte-specific ablation of Stat3 exhibits impaired skin remodeling, but does not affect skin morphogenesis. EMBO J. 18, 4657–4668 (1999).
Chang, H. Y. et al. Gene expression signature of fibroblast serum response predicts human cancer progression: similarities between tumors and wounds. PLOS Biol. 2, E7 (2004).
Fielding, C. A. et al. IL-6 regulates neutrophil trafficking during acute inflammation via STAT3. J. Immunol. 181, 2189–2195 (2008).
Arwert, E. N., Hoste, E. & Watt, F. M. Epithelial stem cells, wound healing and cancer. Nat. Rev. Cancer 12, 170–180 (2012).
Buettner, R. et al. Activated signal transducers and activators of transcription 3 signaling induces CD46 expression and protects human cancer cells from complement-dependent cytotoxicity. Mol. Cancer Res. 5, 823–832 (2007).
Cassetta, L. & Pollard, J. W. Targeting macrophages: therapeutic approaches in cancer. Nat. Rev. Drug Discov. https://doi.org/10.1038/nrd.2018.169 (2018).
Kinchen, J. et al. Structural remodeling of the human colonic mesenchyme in inflammatory bowel disease. Cell 175, 372–386 (2018).
Jarnicki, A., Putoczki, T. & Ernst, M. Stat3: linking inflammation to epithelial cancer - more than a “gut” feeling? Cell Div. 5, 14 (2010).
Medzhitov, R. Origin and physiological roles of inflammation. Nature 454, 428–435 (2008). This review defines para-inflammation as an adaptive and reversible state of subclinical inflammation in response to tissue damage and stress, including tumorigenesis.
Phesse, T. J. et al. Partial inhibition of gp130-Jak-Stat3 signaling prevents Wnt-beta-catenin-mediated intestinal tumor growth and regeneration. Sci. Signal. 7, ra92 (2014). This paper provides genetic evidence that partial suppression of GP130-dependent STAT3 signaling restricts the growth of intestinal tumours arising from excessive activation of canonical WNT signaling, which is a bona fide oncogenic event.
Lasry, A., Zinger, A. & Ben-Neriah, Y. Inflammatory networks underlying colorectal cancer. Nat. Immunol. 17, 230–240 (2016).
Aran, D. et al. Widespread parainflammation in human cancer. Genome Biol. 17, 145 (2016).
Pilati, C. et al. Somatic mutations activating STAT3 in human inflammatory hepatocellular adenomas. J. Exp. Med. 208, 1359–1366 (2011).
Koskela, H. L. et al. Somatic STAT3 mutations in large granular lymphocytic leukemia. N. Engl. J. Med. 366, 1905–1913 (2012).
Barbieri, I. et al. Constitutively active Stat3 enhances neu-mediated migration and metastasis in mammary tumors via upregulation of Cten. Cancer Res. 70, 2558–2567 (2010). This paper genetically clarifies the tumorigenic capacity of a constitutive activating mutation in the endogenous Stat3 allele.
Bromberg, J. F. et al. Stat3 as an oncogene. Cell 98, 295–303 (1999).
Tebbutt, N. C. et al. Reciprocal regulation of gastrointestinal homeostasis by SHP2 and STAT-mediated trefoil gene activation in gp130 mutant mice. Nat. Med. 8, 1089–1097 (2002).
Zenewicz, L. A. et al. IL-22 deficiency alters colonic microbiota to be transmissible and colitogenic. J. Immunol. 190, 5306–5312 (2013).
Schmidt, S. et al. ADAM17 is required for EGF-R-induced intestinal tumors via IL-6 trans-signaling. J. Exp. Med. 215, 1205–1225 (2018).
Grivennikov, S. I. et al. Adenoma-linked barrier defects and microbial products drive IL-23/IL-17-mediated tumour growth. Nature 491, 254–258 (2012).
Putoczki, T. L. et al. Interleukin-11 is the dominant IL-6 family cytokine during gastrointestinal tumorigenesis and can be targeted therapeutically. Cancer Cell 24, 257–271 (2013).
Rajagopal, S., Rajagopal, K. & Lefkowitz, R. J. Teaching old receptors new tricks: biasing seven-transmembrane receptors. Nat. Rev. Drug Discov. 9, 373–386 (2010).
Lee, H. et al. STAT3-induced S1PR1 expression is crucial for persistent STAT3 activation in tumors. Nat. Med. 16, 1421–1428 (2010).
Bollrath, J. et al. gp130-mediated Stat3 activation in enterocytes regulates cell survival and cell-cycle progression during colitis-associated tumorigenesis. Cancer Cell 15, 91–102 (2009).
Garbers, C., Heink, S., Korn, T. & Rose-John, S. Interleukin-6: designing specific therapeutics for a complex cytokine. Nat. Rev. Drug Discov. 17, 395–412 (2018).
Rose-John, S., Winthrop, K. & Calabrese, L. The role of IL-6 in host defence against infections: immunobiology and clinical implications. Nat. Rev. Rheumatol. 13, 399–409 (2017).
Poli, V. & Camporeale, A. STAT3-mediated metabolic reprograming in cellular transformation and implications for drug resistance. Front. Oncol. 5, 121 (2015).
Liang, J. et al. The correlation between the immune and epithelial-mesenchymal transition signatures suggests potential therapeutic targets and prognosis prediction approaches in kidney cancer. Sci. Rep. 8, 6570 (2018).
Marusyk, A. et al. Non-cell-autonomous driving of tumour growth supports sub-clonal heterogeneity. Nature 514, 54–58 (2014).
Bromberg, J. F., Horvath, C. M., Besser, D., Lathem, W. W. & Darnell, J. E. Jr. Stat3 activation is required for cellular transformation by v-src. Mol. Cell. Biol. 18, 2553–2558 (1998).
Kim, D. J., Tremblay, M. L. & Digiovanni, J. Protein tyrosine phosphatases, TC-PTP, SHP1, and SHP2, cooperate in rapid dephosphorylation of Stat3 in keratinocytes following UVB irradiation. PLOS ONE 5, e10290 (2010).
Peyser, N. D. et al. Loss-of-function PTPRD mutations lead to increased STAT3 activation and sensitivity to STAT3 inhibition in head and neck cancer. PLOS ONE 10, e0135750 (2015).
Tartaglia, M. et al. Somatic mutations in PTPN11 in juvenile myelomonocytic leukemia, myelodysplastic syndromes and acute myeloid leukemia. Nat. Genet. 34, 148–150 (2003).
Chung, C. D. et al. Specific inhibition of Stat3 signal transduction by PIAS3. Science 278, 1803–1805 (1997).
Babon, J. J., Varghese, L. N. & Nicola, N. A. Inhibition of IL-6 family cytokines by SOCS3. Semin. Immunol. 26, 13–19 (2014).
El Kasmi, K. C. et al. General nature of the STAT3-activated anti-inflammatory response. J. Immunol. 177, 7880–7888 (2006).
Ip, W. K. E., Hoshi, N., Shouval, D. S., Snapper, S. & Medzhitov, R. Anti-inflammatory effect of IL-10 mediated by metabolic reprogramming of macrophages. Science 356, 513–519 (2017).
Liu, L., McBride, K. M. & Reich, N. C. STAT3 nuclear import is independent of tyrosine phosphorylation and mediated by importin-alpha3. Proc. Natl Acad. Sci. USA 102, 8150–8155 (2005).
Ma, J. & Cao, X. Regulation of Stat3 nuclear import by importin α5 and importin α7 via two different functional sequence elements. Cell. Signal. 18, 1117–1126 (2006).
Aigner, P., Just, V. & Stoiber, D. STAT3 isoforms: alternative fates in cancer? Cytokine https://doi.org/10.1016/j.cyto.2018.07.014 (2018).
Takeda, K. et al. Targeted disruption of the mouse Stat3 gene leads to early embryonic lethality. Proc. Natl Acad. Sci. USA 94, 3801–3804 (1997).
Maritano, D. et al. The STAT3 isoforms alpha and beta have unique and specific functions. Nat. Immunol. 5, 401–409 (2004).
Yoo, J. Y., Huso, D. L., Nathans, D. & Desiderio, S. Specific ablation of Stat3beta distorts the pattern of Stat3-responsive gene expression and impairs recovery from endotoxic shock. Cell 108, 331–344 (2002).
Zhong, Z., Wen, Z. & Darnell, J. E. Jr. Stat3: a STAT family member activated by tyrosine phosphorylation in response to epidermal growth factor and interleukin-6. Science 264, 95–98 (1994).
Wen, Z., Zhong, Z. & Darnell, J. E. Jr. Maximal activation of transcription by Stat1 and Stat3 requires both tyrosine and serine phosphorylation. Cell 82, 241–250 (1995).
Shen, Y. et al. Essential role of STAT3 in postnatal survival and growth revealed by mice lacking STAT3 serine 727 phosphorylation. Mol. Cell. Biol. 24, 407–419 (2004).
Baumgart, S. et al. Inflammation-induced NFATc1-STAT3 transcription complex promotes pancreatic cancer initiation by KrasG12D. Cancer Discov. 4, 688–701 (2014).
Yang, J. et al. Unphosphorylated STAT3 accumulates in response to IL-6 and activates transcription by binding to NFκB. Genes Dev. 21, 1396–1408 (2007).
Zhang, X., Wrzeszczynska, M. H., Horvath, C. M. & Darnell, J. E. Jr. Interacting regions in Stat3 and c-Jun that participate in cooperative transcriptional activation. Mol. Cell. Biol. 19, 7138–7146 (1999).
Schuringa, J. J., Schepers, H., Vellenga, E. & Kruijer, W. Ser727-dependent transcriptional activation by association of p300 with STAT3 upon IL-6 stimulation. FEBS Lett. 495, 71–76 (2001).
Ndubuisi, M. I., Guo, G. G., Fried, V. A., Etlinger, J. D. & Sehgal, P. B. Cellular physiology of STAT3: where’s the cytoplasmic monomer? J. Biol. Chem. 274, 25499–25509 (1999).
Gough, D. J. et al. Mitochondrial STAT3 supports Ras-dependent oncogenic transformation. Science 324, 1713–1716 (2009). This manuscript shows that mitochondrial STAT3 is required for RAS transformation.
Wegrzyn, J. et al. Function of mitochondrial Stat3 in cellular respiration. Science 323, 793–797 (2009). This is the first description of a mitochondrial pool of STAT3 that augments the activity of the ETC.
Avalle, L. et al. STAT3 localizes to the ER, acting as a gatekeeper for ER-mitochondrion Ca2+ fluxes and apoptotic responses. Cell Death Differ. https://doi.org/10.1038/s41418-018-0171-y (2018).
Shah, M. et al. Membrane-associated STAT3 and PY-STAT3 in the cytoplasm. J. Biol. Chem. 281, 7302–7308 (2006).
Shen, S. et al. Cytoplasmic STAT3 represses autophagy by inhibiting PKR activity. Mol. Cell 48, 667–680 (2012). This manuscript shows a direct interaction between STAT3 and PKR that sequesters PKR away from its substrate, eIF2A, and blocks the formation of autophagosomes.
Silver, D. L., Naora, H., Liu, J., Cheng, W. & Montell, D. J. Activated signal transducer and activator of transcription (STAT) 3: localization in focal adhesions and function in ovarian cancer cell motility. Cancer Res. 64, 3550–3558 (2004).
Vander Heiden, M. G., Cantley, L. C. & Thompson, C. B. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science 324, 1029–1033 (2009).
Sabharwal, S. S. & Schumacker, P. T. Mitochondrial ROS in cancer: initiators, amplifiers or an Achilles’ heel? Nat. Rev. Cancer 14, 709–721 (2014).
Demaria, M. et al. A STAT3-mediated metabolic switch is involved in tumour transformation and STAT3 addiction. Aging (Albany NY) 2, 823–842 (2010).
Ziegler, P. K. et al. Mitophagy in intestinal epithelial cells triggers adaptive immunity during tumorigenesis. Cell 174, 88–101 (2018). This study uses preclinical mouse models of colon cancer to show that STAT3 loss results in mitochondrial damage and enhanced mitophagy.
Vassilev, A. O., Lorenz, D. R., Tibbles, H. E. & Uckun, F. M. Role of the leukemia-associated transcription factor STAT3 in platelet physiology. Leuk. Lymphoma 43, 1461–1467 (2002).
Macias, E., Rao, D., Carbajal, S., Kiguchi, K. & DiGiovanni, J. Stat3 binds to mtDNA and regulates mitochondrial gene expression in keratinocytes. J. Invest. Dermatol. 134, 1971–1980 (2014).
Zhang, Q. et al. Mitochondrial localized Stat3 promotes breast cancer growth via phosphorylation of serine 727. J. Biol. Chem. 288, 31280–31288 (2013).
Phillips, D. et al. Stoichiometry of STAT3 and mitochondrial proteins: implications for the regulation of oxidative phosphorylation by protein-protein interactions. J. Biol. Chem. 285, 23532–23536 (2010).
Gough, D. J., Marie, I. J., Lobry, C., Aifantis, I. & Levy, D. E. STAT3 supports experimental K-RasG12D-induced murine myeloproliferative neoplasms dependent on serine phosphorylation. Blood 124, 2252–2261 (2014).
You, L. et al. The role of STAT3 in autophagy. Autophagy 11, 729–739 (2015).
Sargeant, T. J. et al. Stat3 controls cell death during mammary gland involution by regulating uptake of milk fat globules and lysosomal membrane permeabilization. Nat. Cell Biol. 16, 1057–1068 (2014).
Kreuzaler, P. A. et al. Stat3 controls lysosomal-mediated cell death in vivo. Nat. Cell Biol. 13, 303–309 (2011).
Talloczy, Z. et al. Regulation of starvation- and virus-induced autophagy by the eIF2α kinase signaling pathway. Proc. Natl Acad. Sci. USA 99, 190–195 (2002).
Gao, S. P. & Bromberg, J. F. Touched and moved by STAT3. Sci. STKE 2006, pe30 (2006).
Calon, A. et al. Dependency of colorectal cancer on a TGF-β-driven program in stromal cells for metastasis initiation. Cancer Cell 22, 571–584 (2012).
Ng, D. C. et al. Stat3 regulates microtubules by antagonizing the depolymerization activity of stathmin. J. Cell Biol. 172, 245–257 (2006).
Yang, H. et al. STAT3 inhibition enhances the therapeutic efficacy of immunogenic chemotherapy by stimulating type 1 interferon production by cancer cells. Cancer Res. 75, 3812–3822 (2015).
Wolfle, S. J. et al. PD-L1 expression on tolerogenic APCs is controlled by STAT-3. Eur. J. Immunol. 41, 413–424 (2011).
Niu, G. et al. Overexpression of a dominant-negative signal transducer and activator of transcription 3 variant in tumor cells leads to production of soluble factors that induce apoptosis and cell cycle arrest. Cancer Res. 61, 3276–3280 (2001).
Lee, H. et al. A requirement of STAT3 DNA binding precludes Th-1 immunostimulatory gene expression by NF-κB in tumors. Cancer Res. 71, 3772–3780 (2011).
Wynn, T. A. & Barron, L. Macrophages: master regulators of inflammation and fibrosis. Semin. Liver Dis. 30, 245–257 (2010).
Campana, L. et al. The STAT3-IL-10-IL-6 pathway is a novel regulator of macrophage efferocytosis and phenotypic conversion in sterile liver injury. J. Immunol. 200, 1169–1187 (2018).
Gentles, A. J. et al. The prognostic landscape of genes and infiltrating immune cells across human cancers. Nat. Med. 21, 938–945 (2015).
Giurisato, E. et al. Myeloid ERK5 deficiency suppresses tumor growth by blocking protumor macrophage polarization via STAT3 inhibition. Proc. Natl Acad. Sci. USA 115, E2801–E2810 (2018).
Yang, J. et al. Tumor-associated macrophages regulate murine breast cancer stem cells through a novel paracrine EGFR/Stat3/Sox-2 signaling pathway. Stem Cells 31, 248–258 (2013).
Takaishi, K. et al. Involvement of M2-polarized macrophages in the ascites from advanced epithelial ovarian carcinoma in tumor progression via Stat3 activation. Cancer Sci. 101, 2128–2136 (2010).
Takeda, K. et al. Enhanced Th1 activity and development of chronic enterocolitis in mice devoid of Stat3 in macrophages and neutrophils. Immunity 10, 39–49 (1999).
Yan, D., Wang, H. W., Bowman, R. L. & Joyce, J. A. STAT3 and STAT6 signaling pathways synergize to promote cathepsin secretion from macrophages via IRE1α activation. Cell Rep. 16, 2914–2927 (2016).
Lee, Y.-J. et al. Macrophage PD-L1 strikes back: PD-1/PD-L1 interaction drives macrophages toward regulatory subsets. Adv. Biosci. Biotechnol. 4, 35967 (2013).
Mace, T. A. et al. Pancreatic cancer-associated stellate cells promote differentiation of myeloid-derived suppressor cells in a STAT3-dependent manner. Cancer Res. 73, 3007–3018 (2013).
Vasquez-Dunddel, D. et al. STAT3 regulates arginase-I in myeloid-derived suppressor cells from cancer patients. J. Clin. Invest. 123, 1580–1589 (2013). This article uses MDSCs derived from patients with head and neck squamous cell carcinoma to demonstrate that STAT3 directly affects MDSC arginase expression to support the immunosuppressive effects of MDSCs on T cells.
Hossain, D. M. et al. TLR9-targeted STAT3 silencing abrogates immunosuppressive activity of myeloid-derived suppressor cells from prostate cancer patients. Clin. Cancer Res. 21, 3771–3782 (2015).
Dabritz, J., Judd, L. M., Chalinor, H. V., Menheniott, T. R. & Giraud, A. S. Altered gp130 signalling ameliorates experimental colitis via myeloid cell-specific STAT3 activation and myeloid-derived suppressor cells. Sci. Rep. 6, 20584 (2016).
Sumida, K. et al. IL-11 induces differentiation of myeloid-derived suppressor cells through activation of STAT3 signalling pathway. Sci. Rep. 5, 13650 (2015).
Khaled, Y. S., Ammori, B. J. & Elkord, E. Increased levels of granulocytic myeloid-derived suppressor cells in peripheral blood and tumour tissue of pancreatic cancer patients. J. Immunol. Res. 2014, 879897 (2014).
Laouar, Y., Welte, T., Fu, X. Y. & Flavell, R. A. STAT3 is required for Flt3L-dependent dendritic cell differentiation. Immunity 19, 903–912 (2003). This paper identifies STAT3 as a key regulator of FLT3L-mediated development and generation of dendritic cells and their precursor cells.
Flanagan, S. E. et al. Activating germline mutations in STAT3 cause early-onset multi-organ autoimmune disease. Nat. Genet. 46, 812–814 (2014).
Camporeale, A. et al. STAT3 activity is necessary and sufficient for the development of immune-mediated myocarditis in mice and promotes progression to dilated cardiomyopathy. EMBO Mol. Med. 5, 572–590 (2013).
Nieves, E. C. et al. STAT3 expression in host myeloid cells controls graft-versus-host disease severity. Biol. Blood Marrow Transplant. 23, 1622–1630 (2017).
Wang, T. et al. Regulation of the innate and adaptive immune responses by Stat-3 signaling in tumor cells. Nat. Med. 10, 48–54 (2004).
Nefedova, Y. et al. Hyperactivation of STAT3 is involved in abnormal differentiation of dendritic cells in cancer. J. Immunol. 172, 464–474 (2004). This study shows that JAK–STAT3 signalling promotes the generation of immunosuppressive immature dendritic cells.
Gabrilovich, D. Mechanisms and functional significance of tumour-induced dendritic-cell defects. Nat. Rev. Immunol. 4, 941–952 (2004).
Kortylewski, M. et al. Inhibiting Stat3 signaling in the hematopoietic system elicits multicomponent antitumor immunity. Nat. Med. 11, 1314–1321 (2005). This landmark study describes STAT3 as an important signalling node in haematopoietic cells and finds that STAT3 is responsible for suppressing immune-mediated antitumour function in mouse models.
Aden, K. et al. Epithelial IL-23R signaling licenses protective IL-22 responses in intestinal inflammation. Cell Rep. 16, 2208–2218 (2016).
Panopoulos, A. D. et al. STAT3 governs distinct pathways in emergency granulopoiesis and mature neutrophils. Blood 108, 3682–3690 (2006).
Zhang, H. et al. STAT3 controls myeloid progenitor growth during emergency granulopoiesis. Blood 116, 2462–2471 (2010).
Gotthardt, D. et al. Loss of STAT3 in murine NK cells enhances NK cell-dependent tumor surveillance. Blood 124, 2370–2379 (2014).
Jerez, A. et al. STAT3 mutations unify the pathogenesis of chronic lymphoproliferative disorders of NK cells and T cell large granular lymphocyte leukemia. Blood 120, 3048–3057 (2012).
Kucuk, C. et al. Activating mutations of STAT5B and STAT3 in lymphomas derived from gammadelta-T or NK cells. Nat. Commun. 6, 6025 (2015).
Pickert, G. et al. STAT3 links IL-22 signaling in intestinal epithelial cells to mucosal wound healing. J. Exp. Med. 206, 1465–1472 (2009).
Backert, I. et al. STAT3 activation in Th17 and Th22 cells controls IL-22-mediated epithelial host defense during infectious colitis. J. Immunol. 193, 3779–3791 (2014).
Atreya, R. et al. Blockade of interleukin 6 trans signaling suppresses T cell resistance against apoptosis in chronic intestinal inflammation: evidence in crohn disease and experimental colitis in vivo. Nat. Med. 6, 583–588 (2000).
Herrmann, A. et al. CTLA4 aptamer delivers STAT3 siRNA to tumor-associated and malignant T cells. J. Clin. Invest. 124, 2977–2987 (2014).
Yue, C. et al. STAT3 in CD8+T cells inhibits their tumor accumulation by downregulating CXCR3/CXCL10 Axis. Cancer Immunol. Res. 3, 864–870 (2015).
Schmetterer, K. G. et al. STAT3 governs hyporesponsiveness and granzyme B-dependent suppressive capacity in human CD4 + T cells. FASEB J. 29, 759–771 (2015).
Austin, J. W., Lu, P., Majumder, P., Ahmed, R. & Boss, J. M. STAT3, STAT4, NFATc1, and CTCF regulate PD-1 through multiple novel regulatory regions in murine T cells. J. Immunol. 192, 4876–4886 (2014).
Celada, L. J. et al. PD-1 up-regulation on CD4+ T cells promotes pulmonary fibrosis through STAT3-mediated IL-17A and TGF-β1 production. Sci. Transl Med. 10, eaar8356 (2018).
Hsu, P. et al. IL-10 potentiates differentiation of human induced regulatory T cells via STAT3 and Foxo1. J. Immunol. 195, 3665–3674 (2015).
Zorn, E. et al. IL-2 regulates FOXP3 expression in human CD4+CD25+ regulatory T cells through a STAT-dependent mechanism and induces the expansion of these cells in vivo. Blood 108, 1571–1579 (2006).
Kortylewski, M. et al. Regulation of the IL-23 and IL-12 balance by Stat3 signaling in the tumor microenvironment. Cancer Cell 15, 114–123 (2009).
Avery, D. T. et al. B cell-intrinsic signaling through IL-21 receptor and STAT3 is required for establishing long-lived antibody responses in humans. J. Exp. Med. 207, 155–171 (2010).
Meyer-Bahlburg, A. et al. Heterozygous signal transducer and activator of transcription 3 mutations in hyper-IgE syndrome result in altered B cell maturation. J. Allergy Clin. Immunol. 129, 559–562 (2012).
Tangye, S. G., Cook, M. C. & Fulcher, D. A. Insights into the role of STAT3 in human lymphocyte differentiation as revealed by the hyper-IgE syndrome. J. Immunol. 182, 21–28 (2009).
Fornek, J. L. et al. Critical role for Stat3 in T-dependent terminal differentiation of IgG B cells. Blood 107, 1085–1091 (2006).
Herrmann, A. et al. CTLA4 promotes Tyk2-STAT3-dependent B cell oncogenicity. Cancer Res. 77, 5118–5128 (2017).
Zhang, C. et al. CD5 binds to interleukin-6 and induces a feed-forward loop with the transcription factor STAT3 in B cells to promote cancer. Immunity 44, 913–923 (2016).
Kalluri, R. The biology and function of fibroblasts in cancer. Nat. Rev. Cancer 16, 582–598 (2016).
Sanz-Moreno, V. et al. ROCK and JAK1 signaling cooperate to control actomyosin contractility in tumor cells and stroma. Cancer Cell 20, 229–245 (2011).
Laklai, H. et al. Genotype tunes pancreatic ductal adenocarcinoma tissue tension to induce matricellular fibrosis and tumor progression. Nat. Med. 22, 497–505 (2016).
Nagathihalli, N. S. et al. Signal transducer and activator of transcription 3, mediated remodeling of the tumor microenvironment results in enhanced tumor drug delivery in a mouse model of pancreatic cancer. Gastroenterology 149, 1932–1943 (2015).
O’Donoghue, R. J. et al. Genetic partitioning of interleukin-6 signalling in mice dissociates Stat3 from Smad3-mediated lung fibrosis. EMBO Mol. Med. 4, 939–951 (2012).
Augsten, M. Cancer-associated fibroblasts as another polarized cell type of the tumor microenvironment. Front. Oncol. 4, 62 (2014).
Xing, F., Saidou, J. & Watabe, K. Cancer associated fibroblasts (CAFs) in tumor microenvironment. Front. Biosci. (Landmark Ed.) 15, 166–179 (2010).
Albrengues, J. et al. Epigenetic switch drives the conversion of fibroblasts into proinvasive cancer-associated fibroblasts. Nat. Commun. 6, 10204 (2015).
Yang, X. et al. FAP promotes immunosuppression by cancer-associated fibroblasts in the tumor microenvironment via STAT3-CCL2 signaling. Cancer Res. 76, 4124–4135 (2016).
Cheng, Y. et al. Cancer-associated fibroblasts induce PDL1+ neutrophils through the IL6-STAT3 pathway that foster immune suppression in hepatocellular carcinoma. Cell Death Dis. 9, 422 (2018).
Tao, L. et al. Cancer-associated fibroblasts treated with cisplatin facilitates chemoresistance of lung adenocarcinoma through IL-11/IL-11R/STAT3 signaling pathway. Sci. Rep. 6, 38408 (2016).
Yu, H., Kortylewski, M. & Pardoll, D. Crosstalk between cancer and immune cells: role of STAT3 in the tumour microenvironment. Nat. Rev. Immunol. 7, 41–51 (2007).
Liu, Y. et al. STAT3-regulated exosomal miR-21 promotes angiogenesis and is involved in neoplastic processes of transformed human bronchial epithelial cells. Cancer Lett. 370, 125–135 (2016).
Zhao, J. et al. LncRNA PVT1 promotes angiogenesis via activating the STAT3/VEGFA axis in gastric cancer. Oncogene 37, 4094–4109 (2018).
Wu, X. et al. IL-17 promotes tumor angiogenesis through Stat3 pathway mediated upregulation of VEGF in gastric cancer. Tumour Biol. 37, 5493–5501 (2016).
Yan, Q. et al. ANGPTL1 interacts with Integrin α1β1 to suppress HCC angiogenesis and metastasis by inhibiting JAK2/STAT3 signaling. Cancer Res. 77, 5831–5845 (2017).
de la Iglesia, N. et al. Identification of a PTEN-regulated STAT3 brain tumor suppressor pathway. Genes Dev. 22, 449–462 (2008).
Ecker, A. et al. The dark and the bright side of Stat3: proto-oncogene and tumor-suppressor. Front. Biosci. (Landmark Ed) 14, 2944–2958 (2009).
Lee, J. et al. Signal transducer and activator of transcription 3 (STAT3) protein suppresses adenoma-to-carcinoma transition in Apcmin/+ mice via regulation of Snail-1 (SNAI) protein stability. J. Biol. Chem. 287, 18182–18189 (2012).
Musteanu, M. et al. Stat3 is a negative regulator of intestinal tumor progression in Apc(Min) mice. Gastroenterology 138, 1003–1011 (2010).
Couto, J. P. et al. STAT3 negatively regulates thyroid tumorigenesis. Proc. Natl Acad. Sci. USA 109, E2361–E2370 (2012).
Grabner, B. et al. Disruption of STAT3 signalling promotes KRAS-induced lung tumorigenesis. Nat. Commun. 6, 6285 (2015). In KRAS mutant lung adenocarcinoma, lung tissue-specific inactivation of STAT3 in mice results in increased tumour burden, and low STAT3 levels correlate with poor patient survival.
Gao, S. P. et al. Mutations in the EGFR kinase domain mediate STAT3 activation via IL-6 production in human lung adenocarcinomas. J. Clin. Invest. 117, 3846–3856 (2007).
Kopparam, J. et al. RIP4 inhibits STAT3 signaling to sustain lung adenocarcinoma differentiation. Cell Death Differ. 24, 1761–1771 (2017).
Haura, E. B., Zheng, Z., Song, L., Cantor, A. & Bepler, G. Activated epidermal growth factor receptor-Stat-3 signaling promotes tumor survival in vivo in non-small cell lung cancer. Clin. Cancer Res. 11, 8288–8294 (2005).
Brooks, G. D. et al. IL6 trans-signaling promotes KRAS-driven lung carcinogenesis. Cancer Res. 76, 866–876 (2016). This study demonstrates that inhibition of IL-6 trans -signalling ameliorates lung cancer pathogenesis.
D’Amico, S. et al. STAT3 is a master regulator of epithelial identity and KRAS-driven tumorigenesis. Genes Dev. 32, 1175–1187 (2018).
Pencik, J. et al. STAT3 regulated ARF expression suppresses prostate cancer metastasis. Nat. Commun. 6, 7736 (2015). This study shows that the gene encoding ARF is a direct STAT3 target and that genetic ablation of STAT3 signalling disrupts the ARF–MDM2–p53 tumour suppressor axis to bypass mechanisms of cellular senescence in a model of prostate cancer.
Ernst, M. & Putoczki, T. L. Molecular pathways: IL11 as a tumor-promoting cytokine-translational implications for cancers. Clin. Cancer Res. 20, 5579–5588 (2014).
Jones, S. A., Scheller, J. & Rose-John, S. Therapeutic strategies for the clinical blockade of IL-6/gp130 signaling. J. Clin. Invest. 121, 3375–3383 (2011).
Buchert, M., Burns, C. J. & Ernst, M. Targeting JAK kinase in solid tumors: emerging opportunities and challenges. Oncogene 35, 939–951 (2016).
Sakamoto, K. et al. Janus kinase 1 is essential for inflammatory cytokine signaling and mammary gland remodeling. Mol. Cell. Biol. 36, 1673–1690 (2016).
Van Rompaey, L. et al. Preclinical characterization of GLPG0634, a selective inhibitor of JAK1, for the treatment of inflammatory diseases. J. Immunol. 191, 3568–3577 (2013).
Phillips, T. J. et al. Phase 1 study of the PI3Kdelta inhibitor INCB040093+/- JAK1 inhibitor itacitinib in relapsed/refractory B cell lymphoma. Blood 132, 293–306 (2018).
Gadina, M. et al. Translational and clinical advances in JAK-STAT biology: the present and future of jakinibs. J. Leukoc. Biol. 104, 499–514 (2018).
Beebe, J. D., Liu, J. Y. & Zhang, J. T. Two decades of research in discovery of anticancer drugs targeting STAT3, how close are we? Pharmacol. Ther. 191, 74–91 (2018).
Lieu, K. G. et al. The rabies virus interferon antagonist P protein interacts with activated STAT3 and inhibits Gp130 receptor signaling. J. Virol. 87, 8261–8265 (2013).
Zhang, Q. et al. Serum-resistant CpG-STAT3 decoy for targeting survival and immune checkpoint signaling in acute myeloid leukemia. Blood 127, 1687–1700 (2016).
Hong, D. et al. AZD9150, a next-generation antisense oligonucleotide inhibitor of STAT3 with early evidence of clinical activity in lymphoma and lung cancer. Sci. Transl Med. 7, 314ra185 (2015). This paper establishes not only the feasibility of specific targeting of STAT3 in tumour models but also that systemic anti-STAT3 therapy impairs the cell-autonomous processes that are required for the survival and growth of malignant cells as well as limiting their ability to elicit anticancer immune responses.
Kroemer, G., Galluzzi, L. & Zitvogel, L. STAT3 inhibition for cancer therapy: cell-autonomous effects only? Oncoimmunology 5, e1126063 (2016).
Nelson, E. A., Sharma, S. V., Settleman, J. & Frank, D. A. A chemical biology approach to developing STAT inhibitors: molecular strategies for accelerating clinical translation. Oncotarget 2, 518–524 (2011).
Xiang, M. et al. Gene expression-based discovery of atovaquone as a STAT3 inhibitor and anticancer agent. Blood 128, 1845–1853 (2016).
Li, Y. et al. Suppression of cancer relapse and metastasis by inhibiting cancer stemness. Proc. Natl Acad. Sci. USA 112, 1839–1844 (2015).
Locken, H., Clamor, C. & Muller, K. Napabucasin and related heterocycle-fused naphthoquinones as STAT3 inhibitors with antiproliferative activity against cancer cells. J. Nat. Prod. 81, 1636–1644 (2018).
Jonker, D. J. et al. The NCIC CTG and AGITG CO.23 trial: a phase III randomized study of BBI608 plus best supportive care (BSC) versus placebo (PBO) plus BSC in patients (Pts) with pretreated advanced colorectal carcinoma (CRC) [abstract]. J. Clin. Oncol. 32 (Suppl. 15), TPS3660 (2014).
Akira, S. et al. Molecular cloning of APRF, a novel IFN-stimulated gene factor 3 p91-related transcription factor involved in the gp130-mediated signaling pathway. Cell 77, 63–71 (1994).
Alorro, M. G. et al. Generation of an inducible mouse model to reversibly silence Stat3. Genesis 55, e23023 (2017).
Sirkisoon, S. R. et al. Interaction between STAT3 and GLI1/tGLI1 oncogenic transcription factors promotes the aggressiveness of triple-negative breast cancers and HER2-enriched breast cancer. Oncogene 37, 2502–2514 (2018).
Zhang, J. X. et al. Unique genome-wide map of TCF4 and STAT3 targets using ChIP-seq reveals their association with new molecular subtypes of glioblastoma. Neuro Oncol. 15, 279–289 (2013).
Tripathi, S. K. et al. Genome-wide analysis of STAT3-mediated transcription during early human Th17 cell differentiation. Cell Rep. 19, 1888–1901 (2017).
Yang, J. et al. Novel roles of unphosphorylated STAT3 in oncogenesis and transcriptional regulation. Cancer Res. 65, 939–947 (2005).
Yang, J. & Stark, G. R. Roles of unphosphorylated STATs in signaling. Cell Res. 18, 443–451 (2008).
Yuan, Z. L., Guan, Y. J., Chatterjee, D. & Chin, Y. E. Stat3 dimerization regulated by reversible acetylation of a single lysine residue. Science 307, 269–273 (2005).
Lee, J. L., Wang, M. J. & Chen, J. Y. Acetylation and activation of STAT3 mediated by nuclear translocation of CD44. J. Cell Biol. 185, 949–957 (2009).
Nie, Y. et al. STAT3 inhibition of gluconeogenesis is downregulated by SirT1. Nat. Cell Biol. 11, 492–500 (2009).
Limagne, E. et al. Sirtuin-1 activation controls tumor growth by impeding Th17 differentiation via STAT3 deacetylation. Cell Rep. 19, 746–759 (2017).
Dasgupta, M., Dermawan, J. K., Willard, B. & Stark, G. R. STAT3-driven transcription depends upon the dimethylation of K49 by EZH2. Proc. Natl Acad. Sci. USA 112, 3985–3990 (2015).
Yang, J. et al. Reversible methylation of promoter-bound STAT3 by histone-modifying enzymes. Proc. Natl Acad. Sci. USA 107, 21499–21504 (2010).
Kim, E. et al. Phosphorylation of EZH2 activates STAT3 signaling via STAT3 methylation and promotes tumorigenicity of glioblastoma stem-like cells. Cancer Cell 23, 839–852 (2013).
Zhou, Z. et al. SUMOylation and SENP3 regulate STAT3 activation in head and neck cancer. Oncogene 35, 5826–5838 (2016).
Holtick, U., Scheulen, M. E., von Bergwelt-Baildon, M. S. & Weihrauch, M. R. Toll-like receptor 9 agonists as cancer therapeutics. Expert Opin. Investig. Drugs 20, 361–372 (2011).
Redell, M. S., Ruiz, M. J., Alonzo, T. A., Gerbing, R. B. & Tweardy, D. J. Stat3 signaling in acute myeloid leukemia: ligand-dependent and -independent activation and induction of apoptosis by a novel small-molecule Stat3 inhibitor. Blood 117, 5701–5709 (2011).
Xu, X., Kasembeli, M. M., Jiang, X., Tweardy, B. J. & Tweardy, D. J. Chemical probes that competitively and selectively inhibit Stat3 activation. PLOS ONE 4, e4783 (2009).
Horiguchi, A. et al. STAT3 inhibitor WP1066 as a novel therapeutic agent for renal cell carcinoma. Br. J. Cancer 102, 1592–1599 (2010).
Hussain, S. F. et al. A novel small molecule inhibitor of signal transducers and activators of transcription 3 reverses immune tolerance in malignant glioma patients. Cancer Res. 67, 9630–9636 (2007).
Iwamaru, A. et al. A novel inhibitor of the STAT3 pathway induces apoptosis in malignant glioma cells both in vitro and in vivo. Oncogene 26, 2435–2444 (2007).
Sau, S., Mondal, S. K., Kashaw, S. K., Iyer, A. K. & Banerjee, R. Combination of cationic dexamethasone derivative and STAT3 inhibitor (WP1066) for aggressive melanoma: a strategy for repurposing a phase I clinical trial drug. Mol. Cell Biochem. 436, 119–136 (2017).
Chung, S. Y. et al. Two novel SHP-1 agonists, SC-43 and SC-78, are more potent than regorafenib in suppressing the in vitro stemness of human colorectal cancer cells. Cell Death Discov. 5, 25 (2018).
Tai, W. T. et al. Discovery of novel Src homology region 2 domain-containing phosphatase 1 agonists from sorafenib for the treatment of hepatocellular carcinoma. Hepatology 59, 190–201 (2014).
Acknowledgements
This work was supported by grants from the National Health and Medical Research Council (NHMRC) Australia (GNT1079257 and GNT1125951), United States Department of Defense (CA150132), Grant-in-Aid from the Cancer Council Victoria (GNT1145028 and GNT1143036) and funds from the Operational Infrastructure Support Program provided by the Victorian Government. A.C. is a Career Development Fellow (GTN1062247) and M.E. is a Research Fellow of the NHMRC (GNT1079257).
Reviewer information
Nature Reviews Cancer thanks D. Frank and the other anonymous reviewer(s) for their contribution to the peer review of this work.
Author information
Authors and Affiliations
Contributions
All authors researched data for the article, substantially contributed to the discussion of content and wrote, reviewed and edited the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Glossary
- Acute phase response
-
An orchestrated response to tissue injury, infection or inflammation that involves the induction of acute phase proteins by the liver.
- Trans-signalling
-
A process relevant to interleukin-6 (IL-6), and possibly IL-11, whereby enzymatic cleaving of the extracellular domain of the corresponding cognate α-receptors enables formation of soluble ligand–receptor complexes, which can bind to and activate any cell expressing GP130 irrespective of the presence of the native α-receptor chains.
- Para-inflammation
-
An adaptive response to tissue stress or malfunction that is characterized by subclinical reversible inflammation that is manifested by excessive signal transducer and activator of transcription 3 (STAT3) and nuclear factor-κB (NF-κB) activity.
- Electron transport chain
-
(ETC). A series of five protein complexes traversing the mitochondrial inner membrane required for the efficient generation of ATP through oxidative phosphorylation.
- Autophagy
-
A process that is essential to the recycling of unneeded or damaged cellular components during conditions of nutrient stress elicited by many perturbations including cellular transformation and wound healing.
- Autolysosomes
-
Structures that are formed by the fusion of the autophagosome with the lysosome to facilitate digestion of the contents.
Rights and permissions
About this article
Cite this article
Huynh, J., Chand, A., Gough, D. et al. Therapeutically exploiting STAT3 activity in cancer — using tissue repair as a road map. Nat Rev Cancer 19, 82–96 (2019). https://doi.org/10.1038/s41568-018-0090-8
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41568-018-0090-8
This article is cited by
-
Network pharmacology study to explore the multiple molecular mechanism of SH003 in the treatment of non-small cell lung cancer
BMC Complementary Medicine and Therapies (2024)
-
Unraveling the complexity of STAT3 in cancer: molecular understanding and drug discovery
Journal of Experimental & Clinical Cancer Research (2024)
-
IL-8 activates fibroblasts to promote the invasion of HNSCC cells via STAT3-MMP1
Cell Death Discovery (2024)
-
AKT2S128/CCTαS315/319/323-positive cancer-associated fibroblasts (CAFs) mediate focal adhesion kinase (FAK) inhibitors resistance via secreting phosphatidylcholines (PCs)
Signal Transduction and Targeted Therapy (2024)
-
Inhibiting anti-angiogenic VEGF165b activates a miR-17-20a-Calcipressin-3 pathway that revascularizes ischemic muscle in peripheral artery disease
Communications Medicine (2024)