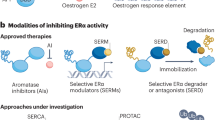
Abstract
Oestrogen receptor-α (ERα), a key driver of breast cancer, normally requires oestrogen for activation. Mutations that constitutively activate ERα without the need for hormone binding are frequently found in endocrine-therapy-resistant breast cancer metastases and are associated with poor patient outcomes. The location of these mutations in the ER ligand-binding domain and their impact on receptor conformation suggest that they subvert distinct mechanisms that normally maintain the low basal state of wild-type ERα in the absence of hormone. Such mutations provide opportunities to probe fundamental issues underlying ligand-mediated control of ERα activity. Instructive contrasts between these ERα mutations and those that arise in the androgen receptor (AR) during anti-androgen treatment of prostate cancer highlight differences in how activation functions in ERs and AR control receptor activity, how hormonal pressures (deprivation versus antagonism) drive the selection of phenotypically different mutants, how altered protein conformations can reduce antagonist potency and how altered ligand–receptor contacts can invert the response that a receptor has to an agonist ligand versus an antagonist ligand. A deeper understanding of how ligand regulation of receptor conformation is linked to receptor function offers a conceptual framework for developing new anti-oestrogens that might be more effective in preventing and treating breast cancer.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Katzenellenbogen, B. S. & Frasor, J. Therapeutic targeting in the estrogen receptor hormonal pathway. Semin. Oncol. 31, 28–38 (2004).
Rugo, H. S. et al. Endocrine therapy for hormone receptor-positive metastatic breast cancer: American Society of Clinical Oncology guideline. J. Clin. Oncol. 34, 3069–3103 (2016).
Tryfonidis, K., Zardavas, D., Katzenellenbogen, B. S. & Piccart, M. Endocrine treatment in breast cancer: cure, resistance and beyond. Cancer Treat. Rev. 50, 68–81 (2016).
Smith, D. F. & Toft, D. O. Minireview: the intersection of steroid receptors with molecular chaperones: observations and questions. Mol. Endocrinol. 22, 2229–2240 (2008).
Pratt, W. B. & Toft, D. O. Regulation of signaling protein function and trafficking by the hsp90/hsp70-based chaperone machinery. Exp. Biol. Med. 228, 111–133 (2003).
Weigelt, B. et al. Molecular portraits and 70-gene prognosis signature are preserved throughout the metastatic process of breast cancer. Cancer Res. 65, 9155–9158 (2005).
The Cancer Genome Atlas Network. Comprehensive molecular portraits of human breast tumours. Nature 490, 61–70 (2012).
Chandarlapaty, S. et al. Prevalence of ESR1 mutations in cell-free DNA and outcomes in metastatic breast cancer: a secondary analysis of the BOLERO-2 clinical trial. JAMA Oncol. 2, 1310–1315 (2016).
Fribbens, C. et al. Plasma ESR1 mutations and the treatment of estrogen receptor-positive advanced breast cancer. J. Clin. Oncol. 34, 2961–2968 (2016).
Spoerke, J. M. et al. Heterogeneity and clinical significance of ESR1 mutations in ER-positive metastatic breast cancer patients receiving fulvestrant. Nat. Commun. 7, 11579 (2016).
Chu, D. et al. ESR1 mutations in circulating plasma tumor DNA from metastatic breast cancer patients. Clin. Cancer. Res. 22, 993–999 (2016).
Fanning, S. W. et al. Estrogen receptor alpha somatic mutations Y537S and D538G confer breast cancer endocrine resistance by stabilizing the activating function-2 binding conformation. eLife 5, e12792 (2016).
Garcia-Murillas, I. et al. Mutation tracking in circulating tumor DNA predicts relapse in early breast cancer. Sci. Transl Med. 7, 302ra133 (2015).
Jeselsohn, R. et al. Emergence of constitutively active estrogen receptor-alpha mutations in pretreated advanced estrogen receptor-positive breast cancer. Clin. Cancer Res. 20, 1757–1767 (2014).
Li, S. et al. Endocrine-therapy-resistant ESR1 variants revealed by genomic characterization of breast-cancer-derived xenografts. Cell Rep. 4, 1116–1130 (2013).
Merenbakh-Lamin, K. et al. D538G mutation in estrogen receptor-alpha: A novel mechanism for acquired endocrine resistance in breast cancer. Cancer Res. 73, 6856–6864 (2013).
Robinson, D. R. et al. Activating ESR1 mutations in hormone-resistant metastatic breast cancer. Nat. Genet. 45, 1446–1451 (2013).
Bahreini, A. et al. Mutation site and context dependent effects of ESR1 mutation in genome-edited breast cancer cell models. Breast Cancer Res. 19, 60 (2017).
Carlson, K. E., Choi, I., Gee, A., Katzenellenbogen, B. S. & Katzenellenbogen, J. A. Altered ligand binding properties and enhanced stability of a constitutively active estrogen receptor: evidence that an open pocket conformation is required for ligand interaction. Biochemistry 36, 14897–14905 (1997).
Clatot, F. et al. Kinetics, prognostic and predictive values of ESR1 circulating mutations in metastatic breast cancer patients progressing on aromatase inhibitor. Oncotarget 7, 74448–74459 (2016).
Mao, C., Livezey, M., Kim, J. E. & Shapiro, D. J. Antiestrogen resistant cell lines expressing estrogen receptor alpha mutations upregulate the unfolded protein response and are killed by BHPI. Scientif. Rep. 6, 34753 (2016).
Toy, W. et al. ESR1 ligand-binding domain mutations in hormone-resistant breast cancer. Nat. Genet. 45, 1439–1445 (2013).
Toy, W. et al. Activating ESR1 mutations differentially affect the efficacy of ER antagonists. Cancer Discov. 7, 277–287 (2017).
Wang, P. et al. Sensitive detection of mono- and polyclonal ESR1 Mutations in primary tumors, metastatic lesions, and cell-free DNA of breast cancer patients. Clin. Cancer Res. 22, 1130–1137 (2016).
Jordan, V. C., Curpan, R. & Maximov, P. Y. Estrogen receptor mutations found in breast cancer metastases integrated with the molecular pharmacology of selective ER modulators. J. Natl Cancer Inst. 107, djv075 (2015).
Pakdel, F., Reese, J. C. & Katzenellenbogen, B. S. Identification of charged residues in an N-terminal portion of the hormone-binding domain of the human estrogen receptor important in transcriptional activity of the receptor. Mol. Endocrinol. 7, 1408–1417 (1993).
Weis, K. E., Ekena, K., Thomas, J. A., Lazennec, G. & Katzenellenbogen, B. S. Constitutively active human estrogen receptors containing amino acid substitutions for tyrosine 537 in the receptor protein. Mol. Endocrinol. 10, 1388–1398 (1996).
Zhang, Q. X., Borg, A., Wolf, D. M., Oesterreich, S. & Fuqua, S. A. An estrogen receptor mutant with strong hormone-independent activity from a metastatic breast cancer. Cancer Res. 57, 1244–1249 (1997).
Joseph, J. D. et al. The selective estrogen receptor downregulator GDC-0810 is efficacious in diverse models of ER+ breast cancer. eLife 5, e15828 (2016).
Nettles, K. W. et al. NFkappaB selectivity of estrogen receptor ligands revealed by comparative crystallographic analyses. Nat. Chem. Biol. 4, 241–247 (2008).
Lazennec, G., Ediger, T. R., Petz, L. N., Nardulli, A. M. & Katzenellenbogen, B. S. Mechanistic aspects of estrogen receptor activation probed with constitutively active estrogen receptors: correlations with DNA and coregulator interactions and receptor conformational changes. Mol. Endocrinol. 11, 1375–1386 (1997).
Kircher, M. & Kelso, J. High-throughput DNA sequencing—concepts and limitations. Bioessays 32, 524–536 (2010).
Zehir, A. et al. Mutational landscape of metastatic cancer revealed from prospective clinical sequencing of 10,000 patients. Nat. Med. 23, 703–713 (2017).
Wilde, O. The Importance of Being Earnest 7th edn (Methuen, 1915).
Trevino, L. S. & Weigel, N. L. Phosphorylation: a fundamental regulator of steroid receptor action. Trends Endocrinol. Metab. 24, 515–524 (2013).
Likhite, V. S., Stossi, F., Kim, K., Katzenellenbogen, B. S. & Katzenellenbogen, J. A. Kinase-specific phosphorylation of the estrogen receptor changes receptor interactions with ligand, deoxyribonucleic acid, and coregulators associated with alterations in estrogen and tamoxifen activity. Mol. Endocrinol. 20, 3120–3132 (2006).
Yee, D. & Lee, A. V. Crosstalk between the insulin-like growth factors and estrogens in breast cancer. J. Mammary Gland Biol. Neoplasia 5, 107–115 (2000).
Voudouri, K., Berdiaki, A., Tzardi, M., Tzanakakis, G. N. & Nikitovic, D. Insulin-like growth factor and epidermal growth factor signaling in breast cancer cell growth: focus on endocrine resistant disease. Anal. Cell Pathol. 2015, 975495 (2015).
Curtis, S. H. & Korach, K. S. Steroid receptor knockout models: phenotypes and responses illustrate interactions between receptor signaling pathways in vivo. Adv. Pharmacol. 47, 357–380 (2000).
Stellato, C. et al. The “busy life” of unliganded estrogen receptors. Proteomics 16, 288–300 (2016).
Iwase, H. Molecular action of the estrogen receptor and hormone dependency in breast cancer. Breast Cancer 10, 89–96 (2003).
Groner, A. C. & Brown, M. Role of steroid receptor and coregulator mutations in hormone-dependent cancers. J. Clin. Invest. 127, 1126–1135 (2017).
Gee, A. C. & Katzenellenbogen, J. A. Probing conformational changes in the estrogen receptor: evidence for a partially unfolded intermediate facilitating ligand binding and release. Mol. Endocrinol. 15, 421–428 (2001).
Seielstad, D. A., Carlson, K. E., Kushner, P. J., Greene, G. L. & Katzenellenbogen, J. A. Analysis of the structural core of the human estrogen receptor ligand binding domain by selective proteolysis/mass spectrometric analysis. Biochemistry 34, 12605–12615 (1995).
White, R., Sjoberg, M., Kalkhoven, E. & Parker, M. G. Ligand-independent activation of the oestrogen receptor by mutation of a conserved tyrosine. EMBO J. 16, 1427–1435 (1997).
Herynk, M. H. & Fuqua, S. A. Estrogen receptor mutations in human disease. Endocr. Rev. 25, 869–898 (2004).
Nwachukwu, J. C. et al. Systems structural biology analysis of ligand effects on ERalpha predicts cellular response to environmental estrogens and anti-hormone therapies. Cell Chem. Biol. 24, 35–45 (2017).
Nwachukwu, J. C. et al. Predictive features of ligand-specific signaling through the estrogen receptor. Mol. Syst. Biol. 12, 864 (2016).
Castoria, G. et al. Tyrosine phosphorylation of estradiol receptor by Src regulates its hormone-dependent nuclear export and cell cycle progression in breast cancer cells. Oncogene 31, 4868–4877 (2012).
Zhao, Y. et al. Structurally novel antiestrogens elicit differential responses from constitutively active mutant estrogen receptors in breast cancer cells and tumors. Cancer Res. 77, 5602–5613 (2017).
Wrenn, C. K. & Katzenellenbogen, B. S. Structure-function analysis of the hormone binding domain of the human estrogen receptor by region-specific mutagenesis and phenotypic screening in yeast. J. Biol. Chem. 268, 24089–24098 (1993).
Madak-Erdogan, Z. et al. Integrative genomics of gene and metabolic regulation by estrogen receptors alpha and beta, and their coregulators. Mol. Syst. Biol. 9, 676 (2013).
Jeyakumar, M., Carlson, K. E., Gunther, J. R. & Katzenellenbogen, J. A. Exploration of dimensions of estrogen potency: parsing ligand binding and coactivator binding affinities. J. Biol. Chem. 286, 12971–12982 (2011).
Zhao, C. et al. Mutation of Leu-536 in human estrogen receptor-alpha alters the coupling between ligand binding, transcription activation, and receptor conformation. J. Biol. Chem. 278, 27278–27286 (2003).
De Mattos-Arruda, L. et al. Capturing intra-tumor genetic heterogeneity by de novo mutation profiling of circulating cell-free tumor DNA: a proof-of-principle. Ann. Oncol. 25, 1729–1735 (2014).
Staby, L. et al. Eukaryotic transcription factors: paradigms of protein intrinsic disorder. Biochem. J. 474, 2509–2532 (2017).
Jain, V. P. & Tu, R. S. Coupled folding and specific binding: fishing for amphiphilicity. Int. J. Mol. Sci. 12, 1431–1450 (2011).
Trizac, E., Levy, Y. & Wolynes, P. G. Capillarity theory for the fly-casting mechanism. Proc. Natl. Acad. Sci. USA 107, 2746–2750 (2010).
Huang, Y. & Liu, Z. Nonnative interactions in coupled folding and binding processes of intrinsically disordered proteins. PLoS ONE 5, e15375 (2010).
Brzozowski, A. M. et al. Molecular basis of agonism and antagonism in the oestrogen receptor. Nature 389, 753–758 (1997).
Shiau, A. K. et al. The structural basis of estrogen receptor/coactivator recognition and the antagonism of this interaction by tamoxifen. Cell 95, 927–937 (1998).
Watson, P. A., Arora, V. K. & Sawyers, C. L. Emerging mechanisms of resistance to androgen receptor inhibitors in prostate cancer. Nat. Rev. Cancer 15, 701–711 (2015).
Lallous, N. et al. Functional analysis of androgen receptor mutations that confer anti-androgen resistance identified in circulating cell-free DNA from prostate cancer patients. Genome. Biol. 17, 10 (2016).
Imamura, Y. & Sadar, M. D. Androgen receptor targeted therapies in castration-resistant prostate cancer: bench to clinic. Int. J. Urol. 23, 654–665 (2016).
Fenton, M. A. et al. Functional characterization of mutant androgen receptors from androgen-independent prostate cancer. Clin. Cancer Res. 3, 1383–1388 (1997).
Veldscholte, J. et al. A mutation in the ligand binding domain of the androgen receptor of human LNCaP cells affects steroid binding characteristics and response to anti-androgens. Biochem. Biophys. Res. Commun. 173, 534–540 (1990).
Yoshida, T. et al. Antiandrogen bicalutamide promotes tumor growth in a novel androgen-dependent prostate cancer xenograft model derived from a bicalutamide-treated patient. Cancer Res. 65, 9611–9616 (2005).
Joseph, J. D. et al. A clinically relevant androgen receptor mutation confers resistance to second-generation antiandrogens enzalutamide and ARN-509. Cancer Discov. 3, 1020–1029 (2013).
Nyquist, M. D. et al. TALEN-engineered AR gene rearrangements reveal endocrine uncoupling of androgen receptor in prostate cancer. Proc. Natl. Acad. Sci. USA 110, 17492–17497 (2013).
Balbas, M. D. et al. Overcoming mutation-based resistance to antiandrogens with rational drug design. eLife 2, e00499 (2013).
McGinley, P. L. & Koh, J. T. Circumventing anti-androgen resistance by molecular design. J. Am. Chem. Soc. 129, 3822–3823 (2007).
Bohl, C. E., Gao, W., Miller, D. D., Bell, C. E. & Dalton, J. T. Structural basis for antagonism and resistance of bicalutamide in prostate cancer. Proc. Natl. Acad. Sci. USA 102, 6201–6206 (2005).
Ince, B. A., Zhuang, Y., Wrenn, C. K., Shapiro, D. J. & Katzenellenbogen, B. S. Powerful dominant negative mutants of the human estrogen receptor. J. Biol. Chem. 268, 14026–14032 (1993).
Ince, B. A., Schodin, D. J., Shapiro, D. J. & Katzenellenbogen, B. S. Repression of endogenous estrogen receptor activity in MCF-7 human breast cancer cells by dominant negative estrogen receptors. Endocrinology 136, 3194–3199 (1995).
Schodin, D. J., Zhuang, Y., Shapiro, D. J. & Katzenellenbogen, B. S. Analysis of mechanisms that determine dominant negative estrogen receptor effectiveness. J. Biol. Chem. 270, 31163–31171 (1995).
Montano, M. M., Ekena, K., Krueger, K. D., Keller, A. L. & Katzenellenbogen, B. S. Human estrogen receptor ligand activity inversion mutants: receptors that interpret antiestrogens as estrogens and estrogens as antiestrogens and discriminate among different antiestrogens. Mol. Endocrinol. 10, 230–242 (1996).
Mahfoudi, A., Roulet, E., Dauvois, S., Parker, M. G. & Wahli, W. Specific mutations in the estrogen receptor change the properties of antiestrogens to full agonists. Proc. Natl. Acad. Sci. USA 92, 4206–4210 (1995).
Wolf, D. M. & Jordan, V. C. The estrogen receptor from a tamoxifen stimulated MCF-7 tumor variant contains a point mutation in the ligand binding domain. Breast Cancer Res. Treatment 31, 129–138 (1994).
Wolf, D. M. & Jordan, V. C. Characterization of tamoxifen stimulated MCF-7 tumor variants grown in athymic mice. Breast Cancer Res. Treatment 31, 117–127 (1994).
Levenson, A. S., Catherino, W. H. & Jordan, V. C. Estrogenic activity is increased for an antiestrogen by a natural mutation of the estrogen receptor. J. Steroid Biochem. Mol. Biol. 60, 261–268 (1997).
Catherino, W. H., Wolf, D. M. & Jordan, V. C. A naturally occurring estrogen receptor mutation results in increased estrogenicity of a tamoxifen analog. Mol. Endocrinol. 9, 1053–1063 (1995).
Levenson, A. S., MacGregor Schafer, J. I., Bentrem, D. J., Pease, K. M. & Jordan, V. C. Control of the estrogen-like actions of the tamoxifen-estrogen receptor complex by the surface amino acid at position 351. J. Steroid Biochem. Mol. Biol. 76, 61–70 (2001).
Bentrem, D. et al. Molecular mechanism of action at estrogen receptor alpha of a new clinically relevant antiestrogen (GW7604) related to tamoxifen. Endocrinology 142, 838–846 (2001).
Liu, H. et al. Structure-function relationships of the raloxifene-estrogen receptor-alpha complex for regulating transforming growth factor-alpha expression in breast cancer cells. J. Biol. Chem. 277, 9189–9198 (2002).
De Savi, C. et al. Optimization of a novel binding motif to (e)-3-(3,5-difluoro-4-((1r,3r)-2-(2-fluoro-2-methylpropyl)-3-methyl-2,3,4,9-tetrahydro-1h-pyrido[3,4-b]indol-1-yl)phenyl)acrylic acid (azd9496), a potent and orally bioavailable selective estrogen receptor downregulator and antagonist. J. Med. Chem. 58, 8128–8140 (2015).
Wu, Y. L. et al. Structural basis for an unexpected mode of SERM-mediated ER antagonism. Mol. Cell 18, 413–424 (2005).
Tora, L. et al. The cloned human oestrogen receptor contains a mutation which alters its hormone binding properties. EMBO J. 8, 1981–1986 (1989).
Levenson, A. S. & Jordan, V. C. Transfection of human estrogen receptor (ER) cDNA into ER-negative mammalian cell lines. J. Steroid Biochem. Mol. Biol. 51, 229–239 (1994).
Levenson, A. S. & Jordan, V. C. The key to the antiestrogenic mechanism of raloxifene is amino acid 351 (aspartate) in the estrogen receptor. Cancer Res. 58, 1872–1875 (1998).
Jiang, S. Y., Parker, C. J. & Jordan, V. C. A model to describe how a point mutation of the estrogen receptor alters the structure-function relationship of antiestrogens. Breast Cancer Res. Treatment 26, 139–147 (1993).
Jiang, S. Y., Langan-Fahey, S. M., Stella, A. L., McCague, R. & Jordan, V. C. Point mutation of estrogen receptor (ER) in the ligand-binding domain changes the pharmacology of antiestrogens in ER-negative breast cancer cells stably expressing complementary DNAs for ER. Mol. Endocrinol. 6, 2167–2174 (1992).
Robertson, J. F. et al. A good drug made better: the fulvestrant dose-response story. Clin. Breast Cancer 14, 381–389 (2014).
Weir, H. M. et al. AZD9496: an oral estrogen receptor inhibitor that blocks the growth of ER-positive and ESR1-mutant breast tumors in preclinical models. Cancer Res. 76, 3307–3318 (2016).
Garner, F., Shomali, M., Paquin, D., Lyttle, C. R. & Hattersley, G. RAD1901: a novel, orally bioavailable selective estrogen receptor degrader that demonstrates antitumor activity in breast cancer xenograft models. Anticancer Drugs 26, 948–956 (2015).
Wardell, S. E. et al. Efficacy of SERD/SERM Hybrid-CDK4/6 inhibitor combinations in models of endocrine therapy-resistant breast cancer Clin. Cancer. Res. 21, 5121–5130 (2015).
Hattersley, G., Harris, A. G., Simon, J. A. & Constantine, G. D. Clinical investigation of RAD1901, a novel estrogen receptor ligand, for the treatment of postmenopausal vasomotor symptoms: a phase 2 randomized, placebo-controlled, double-blind, dose-ranging, proof-of-concept trial. Menopause 24, 92–99 (2017).
Liu, J. et al. Rational design of a boron-modified triphenylethylene (GLL398) as an oral selective estrogen receptor downregulator. ACS Med. Chem. Lett. 8, 102–106 (2017).
Liu, J. et al. Fulvestrant-3 boronic acid (ZB716): an orally bioavailable selective estrogen receptor downregulator (SERD). J. Med. Chem. 59, 8134–8140 (2016).
Jiang, Q., Zhong, Q., Zhang, Q., Zheng, S. & Wang, G. Boron-based 4-hydroxytamoxifen bioisosteres for treatment of de novo tamoxifen resistant breast cancer. ACS Med. Chem. Lett. 3, 392–396 (2012).
Min, J. et al. Adamantyl antiestrogens with novel side chains reveal a spectrum of activities in suppressing estrogen receptor mediated activities in breast cancer cells. J. Med. Chem. 60, 6321–6336 (2017).
Jordan, V. C. Antiestrogens and selective estrogen receptor modulators as multifunctional medicines. 1. Receptor interactions. J. Med. Chem. 46, 883–908 (2003).
Jordan, V. C. Antiestrogens and selective estrogen receptor modulators as multifunctional medicines. 2. Clinical considerations and new agents. J. Med. Chem. 46, 1081–1111 (2003).
Wakeling, A. E. & Bowler, J. ICI 182,780, a new antioestrogen with clinical potential. J. Steroid Biochem. Mol. Biol. 43, 173–177 (1992).
Van de Velde, P. et al. RU 58,668, a new pure antiestrogen inducing a regression of human mammary carcinoma implanted in nude mice. J. Steroid Biochem. Mol. Biol. 48, 187–196 (1994).
Srinivasan, S. et al. Full antagonism of the estrogen receptor without a prototypical ligand side chain. Nat. Chem. Biol. 13, 111–118 (2017).
Zhu, M. et al. Bicyclic core estrogens as full antagonists: synthesis, biological evaluation and structure-activity relationships of estrogen receptor ligands based on bridged oxabicyclic core arylsulfonamides. Org. Biomol. Chem. 10, 8692–8700 (2012).
Zheng, Y. et al. Development of selective estrogen receptor modulator (SERM)-like activity through an indirect mechanism of estrogen receptor antagonism: defining the binding mode of 7-oxabicyclo[2.2.1]hept-5-ene scaffold core ligands. ChemMedChem 7, 1094–1100 (2012).
Lebraud, H. & Heightman, T. D. Protein degradation: a validated therapeutic strategy with exciting prospects. Essays Biochem. 61, 517–527 (2017).
Abdel-Hafiz, H. A. Epigenetic mechanisms of tamoxifen resistance in luminal breast cancer. Diseases 5, E16 (2017).
Nagini, S. Breast cancer: current molecular therapeutic targets and new players. Anticancer Agents Med. Chem. 17, 152–163 (2017).
Song, X. et al. Development of potent small-molecule inhibitors to drug the undruggable steroid receptor coactivator-3. Proc. Natl. Acad. Sci. USA 113, 4970–4975 (2016).
Sahni, J. M. & Keri, R. A. Targeting bromodomain and extraterminal proteins in breast cancer. Pharmacol. Res. https://doi.org/10.1016/j.phrs.2017.11.015 (2017).
Josan, J. S. & Katzenellenbogen, J. A. Designer antiandrogens join the race against drug resistance. eLife 2, e00692 (2013).
Nardone, A., De Angelis, C., Trivedi, M. V., Osborne, C. K. & Schiff, R. The changing role of ER in endocrine resistance. Breast 24 (Suppl. 2), S60–S66 (2015).
Maurer, C., Martel, S., Zardavas, D. & Ignatiadis, M. New agents for endocrine resistance in breast cancer. Breast 34, 1–11 (2017).
Augereau, P. et al. Hormonoresistance in advanced breast cancer: a new revolution in endocrine therapy. Ther. Adv. Med. Oncol. 9, 335–346 (2017).
O’Sullivan, C. C. Overcoming endocrine resistance in hormone-receptor positive advanced breast cancer-the emerging role of CDK4/6 inhibitors. Int. J. Cancer Clin. Res. 2, 029 (2015).
O’Sullivan, C. C. CDK4/6 inhibitors for the treatment of advanced hormone receptor positive breast cancer and beyond: 2016 update. Expert Opin. Pharmacother. 17, 1657–1667 (2016).
Seielstad, D. A., Carlson, K. E., Katzenellenbogen, J. A., Kushner, P. J. & Greene, G. L. Molecular characterization by mass spectrometry of the human estrogen receptor ligand-binding domain expressed in Escherichia coli. Mol. Endocrinol. 9, 647–658 (1995).
Montano, M. M., Müller, V., Trobaugh, A. & Katzenellenbogen, B. S. The carboxy-terminal F domain of the human estrogen receptor: role in the transcriptional activity of the receptor and the effectiveness of antiestrogens as estrogen antagonists. Mol. Endocrinol. 9, 814–825 (1995).
Patel, S. R. & Skafar, D. F. Modulation of nuclear receptor activity by the F domain. Mol. Cell. Endocrinol. 418, 298–305 (2015).
Yang, J., Singleton, D. W., Shaughnessy, E. A. & Khan, S. A. The F-domain of estrogen receptor-alpha inhibits ligand induced receptor dimerization. Mol. Cell. Endocrinol. 295, 94–100 (2008).
Schwartz, J. A., Zhong, L., Deighton-Collins, S., Zhao, C. & Skafar, D. F. Mutations targeted to a predicted helix in the extreme carboxyl-terminal region of the human estrogen receptor-alpha alter its response to estradiol and 4-hydroxytamoxifen. J. Biol. Chem. 277, 13202–13209 (2002).
Tamrazi, A., Carlson, K. E., Daniels, J. R., Hurth, K. M. & Katzenellenbogen, J. A. Estrogen receptor dimerization: ligand binding regulates dimer affinity and dimer dissociation rate. Mol. Endocrinol. 16, 2706–2719 (2002).
Nadal, M. et al. Structure of the homodimeric androgen receptor ligand-binding domain. Nat. Commun. 8, 14388 (2017).
Cheng, Y. & Prusoff, W. H. Relationship between the inhibition constant (K1) and the concentration of inhibitor which causes 50 per cent inhibition (I50) of an enzymatic reaction. Biochem. Pharmacol. 22, 3099–3108 (1973).
Li, Z. et al. Conversion of abiraterone to D4A drives anti-tumour activity in prostate cancer. Nature 523, 347–351 (2015).
Norris, J. D. et al. Androgen receptor antagonism drives cytochrome P450 17A1 inhibitor efficacy in prostate cancer. J. Clin. Invest. 127, 2326–2338 (2017).
Krishnan, A. V. et al. A glucocorticoid-responsive mutant androgen receptor exhibits unique ligand specificity: therapeutic implications for androgen-independent prostate cancer. Endocrinology 143, 1889–1900 (2002).
Acknowledgements
The authors thank their numerous co-workers for their research efforts. Grant support of much of the work described in this Opinion piece from the following sources: the US National Institutes of Health (PHS R01DK015556 to J.A.K., P41GM104601 and T32GM070421 to the University of Illinois, 5R01CA20499 to S.C., P30CA008748 to Memorial Sloan Kettering Cancer Center and P30CA14599 to the University of Chicago Cancer Center), the Virginia and D.K. Ludwig Fund for Cancer Research (to G.L.G.), the US Department of Defense (DOD BC131458 to G.L.G.) and the Breast Cancer Research Foundation (BCRF 17–083 to J.A.K. and B.S.K. and BCRF 17-082 to B.S.K.).
Author information
Authors and Affiliations
Contributions
J.A.K., C.G.M., B.S.K., G.L.G. and S.C. researched the data for the article, provided substantial contributions to discussions of its content, wrote the article and undertook review and/or editing of the manuscript before submission.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Reviewer information
Nature Reviews Cancer thanks J. Carroll, V. C. Jordan and R. Schiff for their contribution to the peer review of this work.
Related links
cBioPortal database: http://www.cbioportal.org/study?id=msk_impact_2017#summary
Electronic supplementary material
41568_2018_1_MOESM2_ESM.mov
Supplementary Movie 1: Activating Mutations in ER – Outside of the ER Ligand Binding Pocket. ERα LBD dimer showing the locations of the activating mutations (green) relative to the ligand (estradiol), the coactivator helix (orange cylinder) and the AF2 coactivator binding region (yellow helices). Monomers are colored blue and gray, and the h9-h10 loop is red. All of the mutations are far from the ligand binding pocket.
41568_2018_1_MOESM3_ESM.mov
Supplementary Movie 2: Activating Mutations in AR – Inside the AR Ligand Binding Pocket. AR LBD monomer showing the locations of the activating mutations (green) relative to the ligand (testosterone), and the AF2 coactivator binding region (yellow helices). All of the AR mutations are inside the ligand binding pocket in contact with the ligand. The extension of h12 can be seen as a kink in the helix, followed by a strand that makes a β-sheet with the h9-h10 loop region and interferes with the dimerization site used by ERα LBD dimers.
41568_2018_1_MOESM4_ESM.mov
Supplementary Movie 3: ER and AR LBDs Overlapped Showing the Locations of the Mutations Relative to the Ligand Binding Pocket. Overlay of ERα LBD (light blue) and the AR LBD (light red) showing the different locations of endocrine therapy resistance mutations (ER – dark blue outside of the LBP, estradiol; AR – dark red, within the LBP). The structure extends beyond the end of h12 in the structure of the AR LBD but not in the ER LBD structure.
Glossary
- Activation functions
-
Regions of amino acid sequence or 3D structure in transcription factors that are associated with the activation of transcription.
- Androgen receptor
-
(AR). A transcription factor that is a member of the nuclear hormone receptor superfamily. It is the principal mediator of the biological effects of androgens and a major driver of the proliferation and progression of prostate cancer.
- Anti-oestrogen
-
A ligand for the oestrogen receptor (ER) used as one form of endocrine therapy for breast cancer. Anti-oestrogens bind to ER and alter its conformation so that it is unable to stimulate the proliferation and progression of breast cancer cells.
- Apo
-
A term that indicates that a binding protein is in its unliganded state.
- Aromatase inhibitors
-
Used as a form of endocrine therapy for breast cancer that works by blocking the production of oestrogens by the ovaries and other tissues, such as the adrenals, and by the tumour itself.
- Conservative mutation
-
The replacement of a residue in a protein with one that has similar physical properties.
- Coulombic repulsion
-
A force separating two entities of equal charge, either positive–positive or negative–negative, when they are close in space.
- Heat shock proteins
-
(HSPs). A family of proteins that selectively bind other proteins that are intrinsically or aberrantly unfolded. HSP90 is the major protein to which wild-type apo-ERα binds, although other HSPs also likely participate in this binding.
- Ligand-binding domain
-
(LBD). A domain of the oestrogen receptor (ER) responsible for binding oestrogens and anti-oestrogens. It is domain E out of the domains A–F and stretches approximately from amino acid 304 to 554 out of a total of 595 amino acids, accounting for about 40% of the overall length of ERα. It is composed of 12 α-helices and a few β-strand elements that make up the secondary structure.
- Ligand-binding pocket
-
(LBP). An interior region of the ligand-binding domain within which both agonist and antagonist ligands bind, with occasional portions of the ligands extending beyond the confines of the pocket.
- Molecular dynamics modelling
-
(MDM). A computationally intensive method for exploring the conformation and dynamic features of proteins by providing alternating inputs of velocity on individual atoms and relaxation within the energy force field confines of the protein.
- Nuclear receptors
-
A superfamily of proteins of which the oestrogen receptor-α (ERα) and the androgen receptor (AR) are members. Most members of the superfamily function largely as transcription factors, many of which are regulated by the binding of ligands, which can be endogenous metabolites (hormones) or exogenous ligands (pharmaceuticals, xenobiotics and so on).
- Oestrogen receptor-α
-
(ERα). A transcription factor that is a member of the nuclear hormone receptor superfamily. The ERα subtype is the principal mediator of the biological effects of oestrogens and a major driver of the proliferation and progression of breast cancer. ERα is distinguished from another ER subtype, ERβ, which has very different biological activities that are largely unrelated to driving breast cancer progression.
- Oestradiol
-
A steroid with an aromatic A ring that is the principal endogenous oestrogen hormone that drives the proliferation and progression of breast cancer cells.
- Selective oestrogen receptor modulator
-
(SERM). A class of oestrogen receptor-α (ERα) ligands that can have tissue-selective pharmacological effects, acting as agonists in some tissues (such as bone and vascular tissues) and antagonists in others (such as breast and uterine tissues). SERMs such as tamoxifen are used in breast cancer endocrine therapy; other SERMs such as raloxifene are used in hormone replacement therapies to protect bone in postmenopausal women.
- Selective oestrogen receptor downregulator
-
(SERD). A class of oestrogen receptor-α (ERα) ligands such as fulvestrant that cause a reduction in the levels of the ERα protein; they also function as ER antagonists and are used in breast cancer endocrine therapies.
Rights and permissions
About this article
Cite this article
Katzenellenbogen, J.A., Mayne, C.G., Katzenellenbogen, B.S. et al. Structural underpinnings of oestrogen receptor mutations in endocrine therapy resistance. Nat Rev Cancer 18, 377–388 (2018). https://doi.org/10.1038/s41568-018-0001-z
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41568-018-0001-z
This article is cited by
-
PSMD14 stabilizes estrogen signaling and facilitates breast cancer progression via deubiquitinating ERα
Oncogene (2024)
-
UTRN as a potential biomarker in breast cancer: a comprehensive bioinformatics and in vitro study
Scientific Reports (2024)
-
Liver tropism of ER mutant breast cancer is characterized by unique molecular changes and immune infiltration
Breast Cancer Research and Treatment (2024)
-
Therapeutic resistance to anti-oestrogen therapy in breast cancer
Nature Reviews Cancer (2023)
-
Clinically relevant CHK1 inhibitors abrogate wild-type and Y537S mutant ERα expression and proliferation in luminal primary and metastatic breast cancer cells
Journal of Experimental & Clinical Cancer Research (2022)