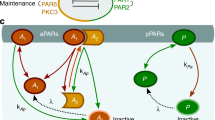
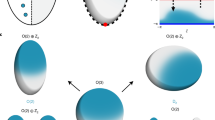
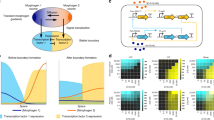
Abstract
Reaction–diffusion networks underlie pattern formation in a range of biological contexts, from morphogenesis of organisms to the polarization of individual cells. One requirement for such molecular networks is that output patterns be scaled to system size. At the same time, kinetic properties of constituent molecules constrain the ability of networks to adapt to size changes. Here, we explore these constraints and the consequences thereof within the conserved PAR cell polarity network. Using the stem-cell-like germ lineage of the Caenorhabditis elegans embryo as a model, we find that the behaviour of PAR proteins fails to scale with cell size. Theoretical analysis demonstrates that this lack of scaling results in a size threshold below which polarity is destabilized, yielding an unpolarized system. In empirically constrained models, this threshold occurs near the size at which germ lineage cells normally switch between asymmetric and symmetric modes of division. Consistent with cell size limiting polarity and division asymmetry, genetic or physical reduction in germ lineage cell size is sufficient to trigger loss of polarity in normally polarizing cells at predicted size thresholds. Physical limits of polarity networks may be one mechanism by which cells read out geometrical features to inform cell fate decisions.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
All data are included in the manuscript or Supplementary material.
Code availability
All model-related code is available at https://github.com/lhcgeneva/PARmodelling. Code for analysis and tracking of particle trajectories is available at https://github.com/lhcgeneva/SPT. Tracking was performed using the trackpy package (https://doi.org/10.5281/zenodo.60550).
References
Rose, L. & Gonczy, P. Polarity establishment, asymmetric division and segregation of fate determinants in early C. elegans embryos. WormBook http://www.wormbook.org/chapters/www_asymcelldiv.2/asymcelldiv.2.html (2014).
Sulston, J., Schierenberg, E., White, J. & Thomson, J. The embryonic cell lineage of the nematode Caenorhabditis elegans. Dev. Biol. 100, 64–119 (1983).
Kemphues, K. J., Priess, J. R., Morton, D. G. & Cheng, N. S. Identification of genes required for cytoplasmic localization in early C. elegans embryos. Cell 52, 311–320 (1988).
Goldstein, B. & Macara, I. G. The PAR proteins: fundamental players in animal cell polarization. Dev. Cell 13, 609–622 (2007).
Goehring, N. W. PAR polarity: from complexity to design principles. Exp. Cell Res. 328, 258–266 (2014).
Motegi, F. et al. Microtubules induce self-organization of polarized PAR domains in Caenorhabditis elegans zygotes. Nat. Cell Biol. 13, 1361–1367 (2011).
Goehring, N. W. et al. Polarization of PAR proteins by advective triggering of a pattern-forming system. Science 334, 1137–1141 (2011).
Gross, P. et al. Guiding self-organized pattern formation in cell polarity establishment. Nat. Phys. 15, 293–300 (2019).
Reich, J. D. et al. Regulated activation of the PAR polarity network ensures a timely and specific response to spatial cues. Curr. Biol. 29, 1911–1923.e5 (2019).
Etemad-Moghadam, B., Guo, S. & Kemphues, K. J. Asymmetrically distributed PAR-3 protein contributes to cell polarity and spindle alignment in early C. elegans embryos. Cell 83, 743–752 (1995).
Watts, J. L. et al. par-6, a gene involved in the establishment of asymmetry in early C. elegans embryos, mediates the asymmetric localization of PAR-3. Development 122, 3133–3140 (1996).
Tabuse, Y. et al. Atypical protein kinase C cooperates with PAR-3 to establish embryonic polarity in Caenorhabditis elegans. Development 125, 3607–3614 (1998).
Gotta, M., Abraham, M. C. & Ahringer, J. CDC-42 controls early cell polarity and spindle orientation in C. elegans. Curr. Biol. 11, 482–488 (2001).
Guo, S. & Kemphues, K. J. par-1, a gene required for establishing polarity in C. elegans embryos, encodes a putative Ser/Thr kinase that is asymmetrically distributed. Cell 81, 611–620 (1995).
Boyd, L., Guo, S., Levitan, D., Stinchcomb, D. T. & Kemphues, K. J. PAR-2 is asymmetrically distributed and promotes association of P granules and PAR-1 with the cortex in C. elegans embryos. Development 122, 3075–3084 (1996).
Hoege, C. et al. LGL can partition the cortex of one-cell Caenorhabditis elegans embryos into two domains. Curr. Biol. 20, 1296–1303 (2010).
Beatty, A., Morton, D. & Kemphues, K. The C. elegans homolog of Drosophila lethal giant larvae functions redundantly with PAR-2 to maintain polarity in the early embryo. Development 137, 3995–4004 (2010).
Kumfer, K. T. et al. CGEF-1 and CHIN-1 regulate CDC-42 activity during asymmetric division in the Caenorhabditis elegans embryo. Mol. Biol. Cell 21, 266–277 (2010).
Tostevin, F. & Howard, M. Modeling the establishment of PAR protein polarity in the one-cell C. elegans embryo. Biophys. J. 95, 4512–4522 (2008).
Dawes, A. T. & Munro, E. M. PAR-3 oligomerization may provide an actin-independent mechanism to maintain distinct par protein domains in the early Caenorhabditis elegans embryo. Biophys. J. 101, 1412–1422 (2011).
Goehring, N. W., Hoege, C., Grill, S. W. & Hyman, A. A. PAR proteins diffuse freely across the anterior-posterior boundary in polarized C. elegans embryos. J. Cell Biol. 193, 583–594 (2011).
Sailer, A., Anneken, A., Li, Y., Lee, S. & Munro, E. Dynamic opposition of clustered proteins stabilizes cortical polarity in the C. elegans zygote. Dev. Cell 35, 131–142 (2015).
Turing, A. M. The chemical basis of morphogenesis. Phil. Trans. R. Soc. Lond. B 237, 37–72 (1952).
Gierer, A. & Meinhardt, H. A theory of biological pattern formation. Kybernetik 12, 30–39 (1972).
Levchenko, A. & Iglesias, P. A. Models of eukaryotic gradient sensing: application to chemotaxis of amoebae and neutrophils. Biophys. J. 82, 50–63 (2002).
Goryachev, A. B. & Pokhilko, A. V. Dynamics of Cdc42 network embodies a Turing-type mechanism of yeast cell polarity. FEBS Lett. 582, 1437–1443 (2008).
Mori, Y., Jilkine, A. & Edelstein-Keshet, L. Wave-pinning and cell polarity from a bistable reaction-diffusion system. Biophys. J. 94, 3684–3697 (2008).
Otsuji, M. et al. A mass conserved reaction-diffusion system captures properties of cell polarity. PLoS Comput. Biol. 3, e108 (2007).
Jilkine, A. & Edelstein-Keshet, L. A comparison of mathematical models for polarization of single eukaryotic cells in response to guided cues. PLoS Comput. Biol. 7, e1001121 (2011).
Halatek, J., Brauns, F. & Frey, E. Self-organization principles of intracellular pattern formation. Phil. Trans. R. Soc. B 373, 20170107 (2018).
Trong, P. K., Nicola, E. M., Goehring, N. W., Kumar, K. V. & Grill, S. W. Parameter-space topology of models for cell polarity. New J. Phys. 16, 065009 (2014).
Rodriguez, J. et al. aPKC cycles between functionally distinct par protein assemblies to drive cell polarity. Dev. Cell 42, 400–415 (2017).
Hara, Y. & Kimura, A. Cell-size-dependent spindle elongation in the Caenorhabditis elegans early embryo. Curr. Biol. 19, 1549–1554 (2009).
Robin, F. B., McFadden, W. M., Yao, B. & Munro, E. M. Single-molecule analysis of cell surface dynamics in Caenorhabditis elegans embryos. Nat. Methods 11, 677–682 (2014).
Schierenberg, E. Reversal of cellular polarity and early cell-cell interaction in the embryo of Caenorhabditis elegans. Dev. Biol. 122, 452–463 (1987).
Homem, C. C. et al. Ecdysone and mediator change energy metabolism to terminate proliferation in Drosophila neural stem cells. Cell 158, 874–888 (2014).
Robinson, S. et al. Generation of spatial patterns through cell polarity switching. Science 333, 1436–1440 (2011).
Roubinet, C. & Cabernard, C. Control of asymmetric cell division. Curr. Opin. Cell Biol. 31, 84–91 (2014).
Grill, S. W., Gönczy, P., Stelzer, E. H. & Hyman, A. A. Polarity controls forces governing asymmetric spindle positioning in the Caenorhabditis elegans embryo. Nature 409, 630–633 (2001).
Ou, G., Stuurman, N., D’Ambrosio, M. & Vale, R. D. Polarized myosin produces unequal-size daughters during asymmetric cell division. Science 330, 677–680 (2010).
Fuse, N., Hisata, K., Katzen, A. L. & Matsuzaki, F. Heterotrimeric G proteins regulate daughter cell size asymmetry in Drosophila neuroblast divisions. Curr. Biol. 13, 947–954 (2003).
Amodeo, A. A. & Skotheim, J. M. Cell-size control. Cold Spring Harb. Perspect. Biol. 8, a019083 (2016).
Thery, M. Micropatterning as a tool to decipher cell morphogenesis and functions. J. Cell Sci. 123, 4201–4213 (2010).
Brenner, S. The genetics of Caenorhabditis elegans. Genetics 77, 71–94 (1974).
Kamath, R. S. et al. Systematic functional analysis of the Caenorhabditis elegans genome using RNAi. Nature 421, 231–237 (2003).
Shelton, C. A. & Bowerman, B. Time-dependent responses to glp-1-mediated inductions in early C. elegans embryos. Development 122, 2043–2050 (1996).
Yamamoto, K. & Kimura, A. An asymmetric attraction model for the diversity and robustness of cell arrangement in nematodes. Development 144, 4437–4449 (2017).
Schenk, C., Bringmann, H., Hyman, A. A. & Cowan, C. R. Cortical domain correction repositions the polarity boundary to match the cytokinesis furrow in C. elegans embryos. Development 137, 1743–1753 (2010).
Schindelin, J. et al. Fiji: an open source platform for biological image analysis. Nat. Methods 9, 676–682 (2012).
Allan, D., Caswell, T., Keim, N. & Van Der Wel, C. trackpy: Trackpy v0.3.2. Zenodo https://doi.org/10.5281/zenodo.60550 (2016).
Dormand, J. & Prince, P. Family of embedded Runge-Kutta formulae. J. Comput. Appl. Math. 6, 19–26 (1980).
Gillespie, D. T. A general method for numerically simulating the stochastic time evolution of coupled chemical reactions. J. Comput. Phys. 22, 403–434 (1976).
Acknowledgements
We thank N. Tapon, B. Baum, R. Endres, C. Weber, J. Pfanzelter, J. Bois and members of the Goehring Lab for critical comments, H. Baumann, B. Atkinson from Intelligent Imaging Innovations (3i), and R. Henriques for providing access to and training for a Marianas LightSheet microscope, the Salbreux Lab for helpful discussions, and T. Hyman and S. Grill, in whose laboratories some of the initial observations were made. This work was supported by the Francis Crick Institute (N.W.G.), which receives its core funding from Cancer Research UK (FC001086), the UK Medical Research Council (FC001086) and the Wellcome Trust (FC001086), the EU Horizon 2020 research and innovation programme under the Marie Skłodowska-Curie grant agreement 675407 (N.W.G.) and a Bogue Fellowship from University College London (L.H.). N.W.G. is a member of the GENiE network supported by COST Action BM1408 and EMBO. We also acknowledge the Santa Barbara Advanced School of Quantitative Biology and the Kavli Institute for Theoretical Physics, supported by NSF grant PHY-1748958, NIH grant R25GM067110 and the Gordon and Betty Moore Foundation grant 2919.01. Some strains were provided by the CGC, which is funded by the NIH Office of Research Infrastructure Programs (P40 OD010440).
Author information
Authors and Affiliations
Contributions
Conceptualization, L.H., N.W.G.; methodology, L.H., F.P., N.T.L.R.; software, L.H.; formal analysis, L.H., N.W.G.; investigation, L.H., F.P., J.D.R., N.H., R.I., N.W.G.; writing, L.H., N.W.G.; funding acquisition, N.W.G.; supervision, N.W.G.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information: Nature Physics thanks James Feng and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary Figs. 1–5 and Tables 1–3.
Supplementary Information
Reporting Summary
Supplementary Video 1
Time evolution of the symmetric PAR model.
Supplementary Video 2
Time-lapse video of an embryo from the zygote stage until division of P4.
Supplementary Video 3
Time-lapse videos capturing P3 division in C27D9.1 (top) and ima-3 (bottom) embryos.
Supplementary Video 4
Time-lapse videos of dissected P0ex (left) and P1ex (right) cells.
Rights and permissions
About this article
Cite this article
Hubatsch, L., Peglion, F., Reich, J.D. et al. A cell-size threshold limits cell polarity and asymmetric division potential. Nat. Phys. 15, 1078–1085 (2019). https://doi.org/10.1038/s41567-019-0601-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41567-019-0601-x
This article is cited by
-
Control of protein-based pattern formation via guiding cues
Nature Reviews Physics (2022)
-
Biophysical Models of PAR Cluster Transport by Cortical Flow in C. elegans Early Embryogenesis
Bulletin of Mathematical Biology (2022)
-
Multilevel regulation of muscle-specific transcription factor hlh-1 during Caenorhabditis elegans embryogenesis
Development Genes and Evolution (2020)
-
Size flips symmetry switch
Nature Physics (2019)