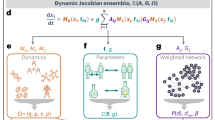
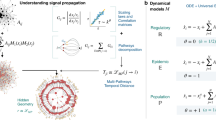
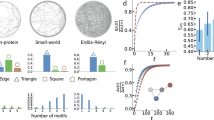
Abstract
A major achievement in the study of complex networks is the realization that diverse systems, from sub-cellular biology to social networks, exhibit universal topological characteristics. Yet, such universality does not naturally translate to the dynamics of these systems, as dynamic behaviour cannot be uniquely predicted from topology alone. Rather, it depends on the interplay of the network’s topology with the dynamic mechanisms of interaction between the nodes. Hence, systems with similar structure may exhibit profoundly different dynamic behaviour. We therefore seek a general theoretical framework to help us systematically translate topological elements into their predicted dynamic outcome. Here, we offer such a translation in the context of signal propagation, linking the topology of a network to the observed spatiotemporal spread of perturbative signals across it, thus capturing the network’s role in propagating local information. For a range of nonlinear dynamic models, we predict that the propagation rules condense into three highly distinctive dynamic regimes, characterized by the interplay between network paths, degree distribution and the interaction dynamics. As a result, classifying a system’s intrinsic interaction mechanisms into the relevant dynamic regime allows us to systematically translate topology into dynamic patterns of information propagation.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
All data and codes to reproduce the results presented here are freely accessible at https://github.com/CRHENS/Spatio-Temporal-/blob/master/README.md. Additional information is available from the corresponding author upon reasonable request. The only exception is the air-traffic network data, which the authors are restricted from sharing.
References
Brockmann, D. & Helbing, D. The hidden geometry of complex, network-driven contagion phenomena. Science 342, 1337–1342 (2013).
Kumar, J., Rotter, S. & Aertsen, A. Spiking activity propagation in neuronal networks: reconciling different perspectives on neural coding. Nat. Rev. Neurosci. 11, 615–627 (2010).
Maslov, S. & Ispolatov, I. Propagation of large concentration changes in reversible protein-binding networks. Proc. Natl Acad. Sci. USA 104, 13655–13660 (2007).
Barzel, B. & Barabási, A.-L. Universality in network dynamics. Nat. Phys. 9, 673–681 (2013).
Barzel, B., Liu, Y.-Y. & Barabási, A.-L. Constructing minimal models for complex system dynamics. Nat. Commun. 6, 7186 (2015).
Holter, N. S., Maritan, A., Cieplak, M., Fedoroff, N. V. & Banavar, J. R. Dynamic modeling of gene expression data. Proc. Natl Acad. Sci. USA 98, 1693–1698 (2001).
Afraimovich, V. S. & Bunimovich, L. A. Dynamical networks: interplay of topology, interactions and local dynamics. Nonlinearity 20, 1761–1771 (2007).
Kirst, C., Timme, M. & Battaglia, D. Dynamic information routing in complex networks. Nat. Commun. 7, 11061 (2016).
Barrat, A., Barthélemy, M. & Vespignani, A. Dynamical Processes on Complex Networks (Cambridge Univ. Press, Cambridge, 2008).
Gao, J., Barzel, B. & Barabási, A.-L. Universal resilience patterns in complex networks. Nature 530, 307–312 (2016).
Harush, U. & Barzel, B. Dynamic patterns of information flow in complex networks. Nat. Commun. 8, 2181 (2017).
Kauffman, S. The ensemble approach to understand genetic regulatory networks. Physica A 340, 733–740 (2004).
Barzel, B. & Biham, O. Quantifying the connectivity of a network: the network correlation function method. Phys. Rev. E 80, 046104 (2009).
Crucitti, P., Latora, V. & Marchiori, M. Model for cascading failures in complex networks. Phys. Rev. E 69, 045104 (2004).
Dobson, I., Carreras, B. A., Lynch, V. E. & Newman, D. E. Complex systems analysis of series of blackouts: cascading failure, critical points, and self-organization. Chaos 17, 026103 (2007).
Voit, E. O. Computational Analysis of Biochemical Systems (Cambridge Univ. Press, New York, NY, 2000).
Alon, U. An Introduction to Systems Biology: Design Principles of Biological Circuits (Chapman & Hall, London, 2006).
Karlebach, G. & Shamir, R. Modelling and analysis of gene regulatory networks. Nat. Rev. 9, 770–780 (2008).
Barabási, A.-L. & Albert, R. Emergence of scaling in random networks. Science 286, 509–512 (1999).
Opsahl, T. & Panzarasa, P. Clustering in weighted networks. Soc. Networks 31, 155–163 (2009).
Eckmann, J.-P., Moses, E. & Sergi, D. Entropy of dialogues creates coherent structures in e-mail traffic. Proc. Natl Acad. Sci. USA 101, 14333–14337 (2004).
Ikehara, K. & Clauset, A. Characterizing the structural diversity of complex networks across domains. Preprint at https://arXiv.org/abs/1710.11304v1 (2017).
Yu, H. et al. High-quality binary protein interaction map of the yeast interactome network. Science 322, 104–110 (2008).
Rual, J. F. et al. Towards a proteome-scale map of the human protein-protein interaction network. Nature 437, 1173–1178 (2005).
Sporns, O., Tononi, G. & Kötter, R. The human connectome: a structural description of the human brain. PLoS Comput. Biol. 1, e42 (2005).
Robertson, C. Flowers and Insects: Lists of Visitors of Four Hundred and Fifty-three Flowers (C. Robertson, Carlinville, Il., 1929).
Pastor-Satorras, R., Castellano, C., Van Mieghem, P. & Vespignani, A. Epidemic processes in complex networks. Rev. Mod. Phys. 87, 925–958 (2015).
Stern, M., Sompolinsky, H. & Abbott, L. F. Dynamics of random neural networks with bistable units. Phys. Rev. E 90, 062710 (2014).
Gardiner, C. W. Handbook of Stochastic Methods (Springer-Verlag, Berlin, 2004).
Castellano, C., Fortunato, S. & Loreto, V. Statistical physics of social dynamics. Rev. Mod. Phys. 81, 591–646 (2009).
Hayes, J. F. & Ganesh Babu, T. V. J. Modeling and Analysis of Telecommunications Networks (John Wiley & Sons, Inc, Hoboken, 2004).
Kuramoto, Y. Chemical Oscillations, Waves and Turbulence (Springer-Verlag, Berlin, Heidelberg, 1984).
Newman, M. E. J. Networks - An Introduction (Oxford Univ. Press, New York, 2010).
Schmetterer, L. & Sigmund, K. (eds) Hans Hahn Gesammelte Abhandlungen Band 1/Hans Hahn Collected Works Vol. 1 (Springer, Vienna, Austria, 1995).
Cohen, R. & Havlin, S. Scale-free networks are ultrasmall. Phys. Rev. Lett. 90, 058701 (2003).
Caldarelli, G. Scale-free Networks: Ccomplex Webs in Nature and Technology (Oxford Univ. Press, New York, 2007).
Wilson, K. G. The renormalization group: critical phenomena and the Kondo problem. Rev. Mod. Phys. 47, 773 (1975).
Bullmore, E. & Sporns, O. Complex brain networks: graph theoretical analysis of structural and functional systems. Nat. Rev. Neurosci. 10, 186–198 (2009).
Wai, H.-T., Scaglione, A., Harush, U., Barzel, B. & Leshem, A. RIDS: robust identification of sparse gene regulatory networks from perturbation experiments. Preprint at https://arxiv.org/abs/1612.06565 (2017).
Novozhilov, A. S., Karev, G. P. & Koonin, E. V. Biological applications of the theory of birth-and-death processes. Brief. Bioinform. 7, 70–85 (2006).
Hufnagel, L., Brockmann, D. & Geisel, T. Forecast and control of epidemics in a globalized world. Proc. Natl Acad. Sci. USA 101, 15124–15129 (2004).
Pastor-Satorras, R. & Vespignani, A. Epidemic spreading in scale-free networks. Phys. Rev. Lett. 86, 3200–3203 (2001).
Dodds, P. S. & Watts, D. J. A generalized model of social and biological contagion. J. Theor. Biol. 232, 587–604 (2005).
Milojević, S. Power-law distributions in information science: making the case for logarithmic binning. J. Am. Soc. Inf. Sci. Technol. 61, 2417–2425 (2010).
Ravaszi, E. B. & Barabási, A.-L. Hierarchical organization in complex networks. Phys. Rev. E 67, 026112 (2003).
Acknowledgements
C.H. thanks the Planning and Budgeting Committee (PBC) of the Council for Higher Education, Israel, and the INSPIRE-Faculty grant (code: IFA17-PH193) for support. This work was supported by the US National Science Foundation-CRISP award no. 1735505, the BIU Center for Research in Applied Cryptography and Cyber Security in conjunction with the Israel National Directorate in the Prime Minister’s office, and by a grant from the Ministry of Science & Technology, Israel & Ministry of Foreign Affairs and International Cooperation General Directorate for Country Promotion, Italian Republic.
Author information
Authors and Affiliations
Contributions
All authors designed and conducted the research. C.H. and U.H. analysed the data and performed the numerical simulations. B.B. was the lead principal investigator.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary Methods, Supplementary Tables 1–5, Supplementary Figures 1–17 and Supplementary References 1–36.
Rights and permissions
About this article
Cite this article
Hens, C., Harush, U., Haber, S. et al. Spatiotemporal signal propagation in complex networks. Nat. Phys. 15, 403–412 (2019). https://doi.org/10.1038/s41567-018-0409-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41567-018-0409-0
This article is cited by
-
Diversity of information pathways drives sparsity in real-world networks
Nature Physics (2024)
-
Uncovering hidden nodes and hidden links in complex dynamic networks
Science China Physics, Mechanics & Astronomy (2024)
-
Sustaining a network by controlling a fraction of nodes
Communications Physics (2023)
-
Diverse electrical responses in a network of fractional-order conductance-based excitable Morris-Lecar systems
Scientific Reports (2023)
-
More is different in real-world multilayer networks
Nature Physics (2023)