Abstract
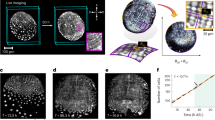
Optimal packings1,2 of unconnected objects have been studied for centuries3,4,5,6, but the packing principles of linked objects, such as topologically complex polymers7,8 or cell lineages9,10, are yet to be fully explored. Here, we identify and investigate a generic class of geometrically frustrated tree packing problems, arising during the initial stages of animal development when interconnected cells assemble within a convex enclosure10. Using a combination of 3D imaging, computational image analysis and mathematical modelling, we study the tree packing problem in Drosophila egg chambers, where 16 germline cells are linked by cytoplasmic bridges to form a branched tree. Our imaging data reveal non-uniformly distributed tree packings, in agreement with predictions from energy-based computations. This departure from uniformity is entropic and affects cell organization during the first stages of the animal’s development. Considering mathematical models of increasing complexity, we investigate spherically confined tree packing problems on convex polyhedra11 that generalize Platonic and Archimedean solids. Our experimental and theoretical results provide a basis for understanding the principles that govern positional ordering in linked multicellular structures, with implications for tissue organization and dynamics12,13.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
Change history
08 August 2018
In the version of this Letter originally published, the citations to equation (1) in Fig. 3 caption and the main text were incorrect; they should have been to equation (2). This has now been corrected.
References
Conway, J. H. & Sloane, N. J. A. Sphere Packings, Lattices, and Groups 3rd edn (Springer, New York, NY, 1999).
Hales, T. C. et al. A revision of the proof of the Kepler conjecture. Discret. Comput. Geom. 44, 1–34 (2010).
Weaire, D. A short history of packing problems. Forma 14, 279–285 (1999).
Donev, A. et al. Improving the density of jammed disordered packings using ellipsoids. Science 303, 990–993 (2004).
Irvine, W. T. M., Vitelli, V. & Chaikin, P. M. Pleats in crystals on curved surfaces. Nature 468, 947–951 (2010).
Meng, G., Paulose, J., Nelson, D. R. & Manoharan, V. N. Elastic instability of a crystal growing on a curved surface. Science 343, 634–637 (2014).
Majikes, J. M., Nash, J. A. & LaBean, T. H. Competitive annealing of multiple DNA origami: formation of chimeric origami. New J. Phys. 18, 115001 (2016).
Uhlmann, F. SMC complexes: from DNA to chromosomes. Nat. Rev. Mol. Cell Biol. 17, 399–412 (2016).
Dayel, M. J. et al. Cell differentiation and morphogenesis in the colony-forming choanoflagellate Salpingoeca rosetta. Dev. Biol. 357, 73–82 (2011).
Haglund, K., Nezis, I. P. & Stenmark, H. Structure and functions of stable intercellular bridges formed by incomplete cytokinesis during development. Commun. Integr. Biol. 4, 1–9 (2011).
Johnson, N. W. Convex solids with regular faces. Can. J. Math. 18, 169–200 (1966).
Hayashi, T. & Carthew, R. W. Surface mechanics mediate pattern formation in the developing retina. Science 431, 647–652 (2004).
Höhn, S., Honerkamp-Smith, A. R., Haas, P. A., Khuc Trong, P. & Goldstein, R. E. Dynamics of a volvox embryo turning itself inside out. Phys. Rev. Lett. 114, 178101 (2015).
Lieberman-Aiden, E. et al. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science 326, 289–293 (2009).
Tomer, R., Khairy, K., Amat, F. & Keller, P. J. Quantitative high-speed imaging of entire developing embryos with simultaneous multiview light-sheet microscopy. Nat. Methods 9, 755–763 (2012).
Ohnesorge, F. & Binnig, G. True atomic resolution by atomic force microscopy through repulsive and attractive forces. Science 260, 1451–1456 (1993).
Manoharan, V. N., Elsesser, M. T. & Pine, D. J. Dense packing and symmetry in small clusters of microspheres. Science 301, 483–487 (2003).
Fletcher, A. J. L. Dry Stone Walls: History and Heritage (Amberley, Stroud, 2016).
Manoharan, V. N. Colloidal matter: packing, geometry, and entropy. Science 349, 1253751 (2015).
Lauga, E. & Brenner, M. P. Evaporation-driven assembly of colloidal particles. Phys. Rev. Lett. 93, 238301 (2004).
Teich, E. G., van Anders, G., Klotsa, D., Dshemuchadse, J. & Glotzer, S. C. Clusters of polyhedra in spherical confinement. Proc. Natl Acad. Sci. USA 113, E669–E678 (2016).
Doig, A. J. & Sternberg, M. J. Side-chain conformational entropy in protein folding. Protein Sci. 4, 2247–2251 (1995).
Glass, D. S. & Riedel-Kruse, I. H. A genetically encoded adhesin toolbox for programming multicellular morphologies and patterns. Preprint at https://doi.org/10.1101/240721 (2017).
Imran Alsous, J., Villoutreix, P., Berezhkovskii, A. M. & Shvartsman, S. Y. Collective growth in a small cell network. Curr. Biol. 27, 2670–2676 (2017).
Gilbert, S. F. Developmental Biology 8th edn (Sinauer Associates, Sunderland, MA, 2006).
Turing, A. M. The chemical basis of morphogenesis. Phil. Trans. R. Soc. B 237, 37–72 (1952).
Cartwright, J. & Arnold, J. M. Intercellular bridges in the embryo of the Atlantic squid, Loligo pealei. Development 57, 3–24 (1980).
Green, K. J., Viamontes, G. L. & Kirk, D. L. Mechanism of formation, ultrastructure, and function of the cytoplasmic bridge system during morphogenesis in Volvox. J. Cell. Biol. 91, 756–769 (1981).
Becalska, A. N. & Gavis, E. R. Lighting up mRNA localization in Drosophila oogenesis. Development 136, 2493–2503 (2009).
Green, K. J. & Kirk, D. L. Cleavage patterns, cell lineages, and development of a cytoplasmic bridge system in Volvox embryos. J. Cell. Biol. 91, 743–755 (1981).
Cromwell, P. R. Polyhedra (Cambridge Univ. Press, Cambridge, 1997).
Thomson, J. J. On the structure of the atom: an investigation of the stability and periods of oscillation of a number of corpuscles arranged at equal intervals around the circumference of a circle; with application of the results to the theory of atomic structure. Philos. Mag. 7, 237–265 (1904).
Allen, M. P. & Tildesley, D. J. Computer Simulation of Liquids (Oxford Univ. Press, Oxford, 1989).
Gould, H. & Tobochnik, J. Statistical and Thermal Physics: With Computer Applications (Princeton Univ. Press, Princeton, NJ, 2010).
Rakhmanov, E. A., Saff, E. B. & Zhou, Y. M. in Computational Methods and Function Theory 1994 (Penang) Vol. 5 (eds Ali, R. M., Ruscheweyh, S. & Saff, E. B.) 293–309 (World Scientific, River Edge, NJ, 1995).
Atiyah, M. & Sutcliffe, P. Polyhedra in physics, chemistry and geometry. Milan J. Math. 71, 33–58 (2003).
Erber, T. & Hockney, G. M. in Advances in Chemical Physics Vol. XCVIII (eds Prigogine, I. & Rice, S. A.) 495–594 (Wiley, New York, NY, 1997).
Acknowledgements
We thank G. Laevsky for expert help with imaging, and E. Gavis, A. Spradling, T. Orr-Weaver, F. Nijhout, L. Manning, H. Mattingly, Y. H. Song, T. Stern, S. Okuda and C. Doherty for helpful discussions. This work was supported by the National Science Foundation Science and Technology Center for Emergent Behaviors of Integrated Cellular Systems CBET-0939511 (J.I.A. and S.Y.S.), the NIH R01GM107103 (S.Y.S.), the WIN programme between Princeton University and the Weizmann Institute (P.V.), a James S. McDonnell Foundation Complex Systems Scholar Award (J.D.) and an Edmund F. Kelly Research Award from the MIT Department of Mathematics (J.D.). This research was partially supported by the Allen Discovery Center programme through The Paul G. Allen Frontiers Group.
Author information
Authors and Affiliations
Contributions
All authors designed the research. J.I.A. performed the experiments. J.I.A., P.V. and N.S. analysed the data. N.S. and J.D. developed the theory. N.S. performed the simulations. All authors wrote the paper.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary Figures S1–S5, Supplementary References 1–14
Rights and permissions
About this article
Cite this article
Imran Alsous, J., Villoutreix, P., Stoop, N. et al. Entropic effects in cell lineage tree packings. Nature Phys 14, 1016–1021 (2018). https://doi.org/10.1038/s41567-018-0202-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41567-018-0202-0
This article is cited by
-
Data-Theoretical Synthesis of the Early Developmental Process
Neuroinformatics (2022)
-
Clonal dominance in excitable cell networks
Nature Physics (2021)