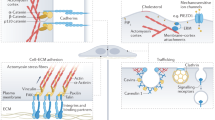
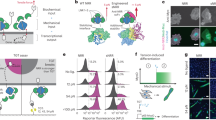
Abstract
Robust and responsive, the surface of a cell is as important as its interior when it comes to mechanically regulating form and function. New techniques are shedding light on this role, and a common language to describe its properties is now needed.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout

Similar content being viewed by others
References
Mayor, R. & Etienne-Manneville, S. The front and rear of collective cell migration. Nat. Rev. Mol. Cell Biol. 17, 97–109 (2016).
Ridley, A. J. Rho GTPase signalling in cell migration. Curr. Opin. Cell Biol. 36, 103–112 (2015).
Raucher, D. & Sheetz, M. P. Cell spreading and lamellipodial extension rate is regulated by membrane tension. J. Cell Biol. 148, 127–136 (2000).
Gauthier, N. C., Fardin, M. A., Roca-Cusachs, P. & Sheetz, M. P. Temporary increase in plasma membrane tension coordinates the activation of exocytosis and contraction during cell spreading. Proc. Natl Acad. Sci. USA 108, 11467–11472 (2011).
Keren, K. et al. Mechanism of shape determination in motile cells. Nature 453, 475–480 (2008).
Mitchison, T. & Cramer, L. Actin-based cell motility and cell locomotion. Cell 84, 371–379 (1996).
Houk, A. R. et al. Membrane tension maintains cell polarity by confining signals to the leading edge during neutrophil migration. Cell 148, 175–188 (2012).
Kozlov, M. M. & Mogilner, A. Model of polarization and bistability of cell fragments. Biophys. J. 93, 3811–3819 (2007).
Diz-Muñoz, A. et al. Membrane tension acts through PLD2 and mTORC2 to limit actin network assembly during neutrophil migration. PLoS Biol. 14, e1002474 (2016).
Tsujita, K., Takenawa, T. & Itoh, T. Feedback regulation between plasma membrane tension and membrane-bending proteins organizes cell polarity during leading edge formation. Nat. Cell Biol. 17, 749–758 (2015).
Lieber, A. D., Schweitzer, Y., Kozlov, M. M. & Keren, K. Front-to-rear membrane tension gradient in rapidly moving cells. Biophys. J. 108, 1599–1603 (2015).
Iskratsch, T., Wolfenson, H. & Sheetz, M. P. Appreciating force and shape — the rise of mechanotransduction in cell biology. Nat. Rev. Mol. Cell Biol. 15, 825–833 (2014).
Atilla-Gokcumen, G. E. et al. Dividing cells regulate their lipid composition and localization. Cell 156, 428–439 (2014).
Dai, J. & Sheetz, M. P. Membrane tether formation from blebbing cells. Biophys. J. 77, 3363–3370 (1999).
Hayashi, K., Yonemura, S., Matsui, T. & Tsukita, S. Immunofluorescence detection of ezrin/radixin/moesin (ERM) proteins with their carboxyl-terminal threonine phosphorylated in cultured cells and tissues. J. Cell Sci. 112, 1149–1158 (1999).
Link, V. et al. Identification of regulators of germ layer morphogenesis using proteomics in zebrafish. J. Cell Sci. 119, 2073–2083 (2006).
Bennett, V. & Baines, A. J. Spectrin and ankyrin-based pathways: metazoan inventions for integrating cells into tissues. Physiol. Rev. 81, 1353–1392 (2001).
Krieg, M., Dunn, A. R. & Goodman, M. B. Mechanical control of the sense of touch by β-spectrin. Nat. Cell Biol. 16, 224–233 (2014).
Gaetani, M., Mootien, S., Harper, S., Gallagher, P. G. & Speicher, D. W. Structural and functional effects of hereditary hemolytic anemia-associated point mutations in the alpha spectrin tetramer site. Blood 111, 5712–5720 (2008).
Ikeda, Y. et al. Spectrin mutations cause spinocerebellar ataxia type 5. Nat. Genet. 38, 184–190 (2006).
Simunovic, M. et al. Friction mediates scission of tubular membranes scaffolded by BAR proteins. Cell 170, 172–184 (2017).
Lewis, A. H. & Grandl, J. Mechanical sensitivity of Piezo1 ion channels can be tuned by cellular membrane tension. eLife 4, (2015).
Evans, E. A. Bending elastic modulus of red blood cell membrane derived from buckling instability in micropipet aspiration tests. Biophys. J. 43, 27–30 (1983).
Hochmuth, F. M., Shao, J. Y., Dai, J. & Sheetz, M. P. Deformation and flow of membrane into tethers extracted from neuronal growth cones. Biophys. J. 70, 358–369 (1996).
Charras, G. & Paluch, E. Blebs lead the way: how to migrate without lamellipodia. Nat. Rev. Mol. Cell Biol. 9, 730–736 (2008).
Sens, P. & Plastino, J. Membrane tension and cytoskeleton organization in cell motility. J. Phys. Condens. Matter 27, 273103 (2015).
Sheetz, M. Cell control by membrane-cytoskeleton adhesion. Nat. Rev. Mol. Cell Biol. 2, 392–396 (2001).
Sun, M. et al. Multiple membrane tethers probed by atomic force microscopy. Biophys. J. 89, 4320–4329 (2005).
Sun, M. et al. The effect of cellular cholesterol on membrane-cytoskeleton adhesion. J. Cell Sci. 120, 2223–2231 (2007).
Brochard-Wyart, F., Borghi, N., Cuvelier, D. & Nassoy, P. Hydrodynamic narrowing of tubes extruded from cells. Proc. Natl Acad. Sci. USA 103, 7660–7663 (2006).
Waugh, R. & Evans, E. A. Thermoelasticity of red blood cell membrane. Biophys. J. 26, 115–131 (1979).
Dimova, R. Advances in Colloid and Interface Science. Adv. Colloid Interface Sci. 208, 225–234 (2014).
Salbreux, G., Charras, G. & Paluch, E. Actin cortex mechanics and cellular morphogenesis. Trends Cell Biol. 22, 536–545 (2012).
Chugh, P. et al. Actin cortex architecture regulates cell surface tension. Nat. Cell Biol. 19, 689–697 (2017).
Bovellan, M. et al. Cellular control of cortical actin nucleation. Curr. Biol. 24, 1628–1635 (2014).
Fritzsche, M., Erlenka mper, C., Moeendarbary, E., Charras, G. & Kruse, K. Actin kinetics shapes cortical network structure and mechanics. Sci. Adv. 2, e1501337 (2016).
Suarez, C. & Kovar, D. R. Internetwork competition for monomers governs actin cytoskeleton organization. Nat. Rev. Mol. Cell Biol. 17, 799–810 (2016).
Evans, E. & Yeung, A. Apparent viscosity and cortical tension of blood granulocytes determined by micropipet aspiration. Biophys. J. 56, 151–160 (1989).
Dai, J., Ting-Beall, H. P., Hochmuth, R. M., Sheetz, M. P. & Titus, M. A. Myosin I contributes to the generation of resting cortical tension. Biophys. J. 77, 1168–1176 (1999).
Schwarz, E. C., Neuhaus, E. M., Kistler, C., Henkel, A. W. & Soldati, T. Dictyostelium myosin IK is involved in the maintenance of cortical tension and affects motility and phagocytosis. J. Cell Sci. 113, 621–633 (2000).
Ledesma, M. D. & Dotti, C. G. Membrane and cytoskeleton dynamics during axonal elongationand stabilization. Int. Rev. Cytol. 227, 183–219 (2003).
Diz-Muñoz, A. et al. Control of directed cell migration in vivo by membrane-to-cortex attachment. PLoS Biol. 8, e1000544 (2010).
Bieling, P. et al. Force feedback controls motor activity and mechanical properties of self-assembling branched actin networks. Cell 164, 115–127 (2016).
Betz, T., Lenz, M., Joanny, J.-F. & Sykes, C. ATP-dependent mechanics of red blood cells. Proc. Natl Acad. Sci. USA 106, 15320–15325 (2009).
Pontes, B. et al. Membrane tension controls adhesion positioning at the leading edge of cells. J. Cell Biol. 216, 2959 (2017).
Schmidtke, D. W. & Diamond, S. L. Direct observation of membrane tethers formed during neutrophil attachment to platelets or P-selectin under physiological flow. J. Cell Biol. 149, 719–730 (2000).
Fischer-Friedrich, E., Hyman, A. A., Jülicher, F., Muller, D. J. & Helenius, J. Quantification of surface tension and internal pressure generated by single mitotic cells. Sci. Rep. 4, 137 (2014).
Yoneda, M. & Dan, K. Tension at the surface of the dividing sea-urchin egg. J. Exp. Biol. 57, 575–587 (1972).
Mayer, M., Depken, M., Bois, J. S., Jülicher, F. & Grill, S. W. Anisotropies in cortical tension reveal the physical basis of polarizing cortical flows. Nature 467, 617–621 (2010).
Rosenbluth, M. J., Lam, W. A. & Fletcher, D. A. Force microscopy of nonadherent cells: a comparison of leukemia cell deformability. Biophys. J. 90, 2994–3003 (2006).
Elsayad, K. et al. Mapping the subcellular mechanical properties of live cells in tissues with fluorescence emission-Brillouin imaging. Sci. Signal. 9, rs5 (2016).
Antonacci, G. & Braakman, S. Biomechanics of subcellularstructures by non-invasive Brillouin microscopy. Sci. Rep. 6, 37217 (2016).
Coughlin, M. F. et al. Cytoskeletal stiffness, friction, and fluidity of cancer cell lines with different metastatic potential. Clin. Exp. Metastasis 30, 237–250 (2012).
Hanakam, F., Albrecht, R., Eckerskorn, C., Matzner, M. & Gerisch, G. Myristoylated and non-myristoylated forms of the pH sensor protein hisactophilin II: intracellular shuttling to plasma membrane and nucleus monitored in real time by a fusion with green fluorescent protein. EMBO J. 15, 2935–2943 (1996).
Clark, A. G., Dierkes, K. & Paluch, E. K. Monitoring actin cortex thickness in live cells. Biophys. J. 105, 570–580 (2013).
Zhelev, D. V., Needham, D. & Hochmuth, R. M. Role of the membrane cortex in neutrophil deformation in small pipets. Biophys. J. 67, 696–705 (1994).
Fricke, K., Wirthensohn, K., Laxhuber, R. & Sackmann, E. Flicker spectroscopy of erythrocytes. A sensitive method to study subtle changes of membrane bending stiffness. Eur. Biophys. J. 14, 67–81 (1986).
López-Duarte, I., Vu, T. T., Izquierdo, M. A., Bull, J. A. & Kuimova, M. K. A molecular rotor for measuring viscosity in plasma membranes of live cells. Chem. Commun. 50, 5282–5284 (2014).
Campàs, O. A toolbox to explore the mechanics of living embryonic tissues. Semin. Cell Dev. Biol. 55, 119–130 (2016).
Sugimura, K., Lenne, P. F. & Graner, F. Measuring forces and stresses in situ in living tissues. Development 143, 186–196 (2016).
Polacheck, W. J. & Chen, C. S. Measuring cell-generated forces: a guide to the available tools. Nat. Methods 13, 415–423 (2016).
Roca-Cusachs, P., Conte, V. & Trepat, X. Quantifying forces in cell biology. Nat. Cell Biol. 19, 742–751 (2017).
Acknowledgements
We thank M. Bergert for comments on the manuscript and the schematics in Fig. 1. We acknowledge the financial support of the European Molecular Biology Laboratory (A.D.-M.), the NIH through GM114671 (D.A.F.), GM114344 (D.A.F) and GM118167 (O.D.W.), and the Chan Zuckerberg Biohub (D.A.F.).
Author information
Authors and Affiliations
Corresponding authors
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Diz-Muñoz, A., Weiner, O.D. & Fletcher, D.A. In pursuit of the mechanics that shape cell surfaces. Nature Phys 14, 648–652 (2018). https://doi.org/10.1038/s41567-018-0187-8
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41567-018-0187-8
This article is cited by
-
The C. elegans homolog of human panic-disorder risk gene TMEM132D orchestrates neuronal morphogenesis through the WAVE-regulatory complex
Molecular Brain (2021)
-
Homeostatic membrane tension constrains cancer cell dissemination by counteracting BAR protein assembly
Nature Communications (2021)
-
Kindlin3 regulates biophysical properties and mechanics of membrane to cortex attachment
Cellular and Molecular Life Sciences (2021)
-
High-throughput cell and spheroid mechanics in virtual fluidic channels
Nature Communications (2020)
-
Effects of energy metabolism on the mechanical properties of breast cancer cells
Communications Biology (2020)