Abstract
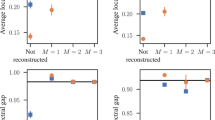
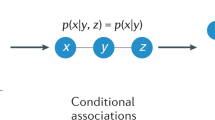
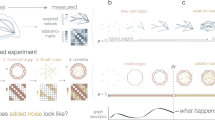
Driven by growing interest across the sciences, a large number of empirical studies have been conducted in recent years of the structure of networks ranging from the Internet and the World Wide Web to biological networks and social networks. The data produced by these experiments are often rich and multimodal, yet at the same time they may contain substantial measurement error1,2,3,4,5,6,7. Accurate analysis and understanding of networked systems requires a way of estimating the true structure of networks from such rich but noisy data8,9,10,11,12,13,14,15. Here we describe a technique that allows us to make optimal estimates of network structure from complex data in arbitrary formats, including cases where there may be measurements of many different types, repeated observations, contradictory observations, annotations or metadata, or missing data. We give example applications to two different social networks, one derived from face-to-face interactions and one from self-reported friendships.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout

Similar content being viewed by others
References
Killworth, P. D. & Bernard, H. R. Informant accuracy in social network data. Hum. Organ. 35, 269–286 (1976).
Marsden, P. V. Network data and measurement. Annu. Rev. Sociol. 16, 435–463 (1990).
Lakhina, A., Byers, J., Crovella, M. & Xie, P. Sampling biases in IP topology measurements. In Proc. 22nd Annual Joint Conf. of the IEEE Computer and Communications Societies (Institute of Electrical and Electronics Engineers, New York, NY, 2003).
Clauset, A. & Moore, C. Accuracy and scaling phenomena in Internet mapping. Phys. Rev. Lett. 94, 018701 (2005).
Wodak, S. J., Pu, S., Vlasblom, J. & Séraphin, B. Challenges and rewards of interaction proteomics. Mol. Cell. Proteom. 8, 3–18 (2009).
Handcock, M. S. & Gile, K. J. Modeling social networks from sampled data. Ann. Appl. Stat. 4, 5–25 (2010).
Lusher, D., Koskinen, J. & Robins, G. Exponential Random Graph Models for Social Networks: Theory, Methods, and Applications (Cambridge Univ. Press, Cambridge, 2012).
Butts, C. T. Network inference, error, and informant (in)accuracy: A Bayesian approach. Soc. Netw. 25, 103–140 (2003).
Clauset, A., Moore, C. & Newman, M. E. J. Hierarchical structure and the prediction of missing links in networks. Nature 453, 98–101 (2008).
Guimerà, R. & Sales-Pardo, M. Missing and spurious interactions and the reconstruction of complex networks. Proc. Natl Acad. Sci. USA 106, 22073–22078 (2009).
Namata, G. M., Kok, S. & Getoor, L. Collective graph identification. In Proc. 17th ACM SIGKDD International Conference on Knowledge Discovery and Data Mining (Association of Computing Machinery, New York, 2011).
Allen, J. D., Xie, Y., Chen, M., Girard, L. & Xiao, G. Comparing statistical methods for constructing large scale gene networks. PLoS One 7, e29348 (2012).
Han, X., Shen, Z., Wang, W.-X. & Di, Z. Robust reconstruction of complex networks from sparse data. Phys. Rev. Lett. 114, 028701 (2015).
Martin, T., Ball, B. & Newman, M. E. J. Structural inference for uncertain networks. Phys. Rev. E 93, 012306 (2016).
Casiraghi, G., Nanumyan, V., Scholtes, I. & Schweitzer, F. From relational data to graphs: Inferring significant links using generalized hypergeometric ensembles. In Proc. International Conf. on Social Informatics (SocInfo 2017), no. 10540 in Lecture Notes in Computer Science (eds Ciampaglia, G. et al.) 111–120 (Springer, Berlin, 2017).
Uetz, P. et al. A comprehensive analysis of protein–protein interactions in Saccharomyces cerevisiae. Nature 403, 623–627 (2000).
Ito, T. et al. A comprehensive two-hybrid analysis to explore the yeast protein interactome. Proc. Natl Acad. Sci. USA 98, 4569–4574 (2001).
Giot, L., Bader, J. S. & Brouwer, C. et al. A protein interaction map of Drosophila melanogaster. Science 302, 1727–1736 (2003).
Krogan, N. J. et al. Global landscape of protein complexes in the yeast Saccharomyces cerevisiae. Nature 440, 637–643 (2006).
Rapoport, A. & Horvath, W. J. A study of a large sociogram. Behav. Sci. 6, 279–291 (1961).
Resnick, M. D. et al. Protecting adolescents from harm: Findings from the National Longitudinal Study on Adolescent Health. J. Am. Med. Assoc. 278, 823–832 (1997).
Bernard, H. R. & Killworth, P. D. Informant accuracy in social network data II. Human. Commun. Res. 4, 3–18 (1977).
Liu, Y., Liu, N. J. & Zhao, H. Y. Inferring protein–protein interactions through high-throughput interaction data from diverse organisms. Bioinformatics 21, 3279–3285 (2005).
Angulo, M. T., Moreno, J. A., Lippner, G., Barabási, A.-L. & Liu, Y.-Y. Fundamental limitations of network reconstruction from temporal data. J. Royal Soc. Interface 14, 20160966 (2017).
Overbeek, R. et al. Wit: Integrated system for high-throughput genome sequence analysis and metabolic reconstruction. Nucleic Acids Res. 28, 123–125 (2000).
Forster, J., Famili, I., Fu, P., Palsson, B. O. & Nielsen, J. Genome-scale reconstruction of the Saccharomyces cerevisiae metabolic network. Genome Res. 13, 244–253 (2003).
Schafer, J. & Strimmer, K. An empirical Bayes approach to inferring large-scale gene association networks. Bioinformatics 21, 754–764 (2005).
Margolin, A. A. et al. ARACNE: An algorithm for the reconstruction of gene regulatory networks in a mammalian cellular context. BMC Bioinformatics 7, S7 (2006).
Langfelder, P. & Horvath, S. Wgcna: An R package for weighted correlation network analysis. BMC Bioinformatics 9, 559 (2008).
Liben-Nowell, D. & Kleinberg, J. The link-prediction problem for social networks. J. Assoc. Inf. Sci. Technol. 58, 1019–1031 (2007).
Huisman, M. Imputation of missing network data: Some simple procedures. J. Social Struct. 10, 1–29 (2009).
Kim, M. & Leskovec, J. The network completion problem: Inferring missing nodes and edges in networks. In Proc. 2011 SIAM International Conf. on Data Mining (eds Liu, B. et al.) 47–58 (Society for Industrial and Applied Mathematics: Philadelphia, PA, 2011).
Smalheiser, N. R. & Torvik, V. I. Author name disambiguation. Annu. Rev. Inf. Sci. Technol. 43, 287–313 (2009).
D’Angelo, C. A., Giuffrida, C. & Abramo, G. A heuristic approach to author name disambiguation in bibliometrics databases for large-scale research assessments. J. Assoc. Inf. Sci. Technol. 62, 257–269 (2011).
Ferreira, A. A., Goncalves, M. A. & Laender, A. H. F. A brief survey of automatic methods for author name disambiguation. SIGMOD Rec. 41, 15–26 (2012).
Tang, J., Fong, A. C. M., Wang, B. & Zhang, J. A unified probabilistic framework for name disambiguation in digital library. IEEE Trans. Knowl. Data Eng. 24, 975–987 (2012).
Brugere, I., Gallagher, B. & Berger-Wolf, T. Y. Network structure inference, a survey: Motivations, methods, and applications. ACM Comput. Surv. 1, 1 (2016).
Dempster, A. P., Laird, N. M. & Rubin, D. B. Maximum likelihood from incomplete data via the EM algorithm. J. Royal Stat. Soc. B 39, 185–197 (1977).
Eagle, N. & Pentland, A. Reality mining: Sensing complex social systems. J. Personal Ubiquitous Comput. 10, 255–268 (2006).
Acknowledgements
The author thanks E. Bruch, G. Cantwell, T. Martin, G. Reinert and M. Riolofor useful comments. This work was funded in part by the US National Science Foundation under grants DMS–1407207 and DMS–1710848. This work uses data from Add Health, a programme project designed by J. R. Udry, P. S. Bearman and K. Mullan Harris, and funded by a grant P01–HD31921 from the Eunice Kennedy Shriver National Institute of Child Health and Human Development, with cooperative funding from 23 other federal agencies and foundations. A special acknowledgment is due to R. R. Rindfuss and B. Entwisle for assistance in the original design. Anyone interested in obtaining data files from Add Health should contact Add Health, Carolina Population Center, 123 W. Franklin Street, Chapel Hill, NC 27516-2524 (addhealth@unc.edu). No direct support was received from grant P01-HD31921 for this analysis.
Author information
Authors and Affiliations
Contributions
M.E.J.N. designed and conducted the research and wrote the paper.
Corresponding author
Ethics declarations
Competing interests
The author declares no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Material
Supplementary notes, supplementary figures 1–3
Rights and permissions
About this article
Cite this article
Newman, M.E.J. Network structure from rich but noisy data. Nature Phys 14, 542–545 (2018). https://doi.org/10.1038/s41567-018-0076-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41567-018-0076-1
This article is cited by
-
Understanding the complexities of Bluetooth for representing real-life social networks
Personal and Ubiquitous Computing (2024)
-
Link prediction using deep autoencoder-like non-negative matrix factorization with L21-norm
Applied Intelligence (2024)
-
Compressing network populations with modal networks reveal structural diversity
Communications Physics (2023)
-
Reconstructing signed relations from interaction data
Scientific Reports (2023)
-
Hypergraph reconstruction from uncertain pairwise observations
Scientific Reports (2023)