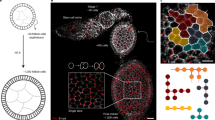
Abstract
The emergence of complex organs is driven by the coordinated proliferation, migration and differentiation of precursor cells. The fate behaviour of these cells is reflected in the time evolution of their progeny, termed clones, which serve as a key experimental observable. In adult tissues, where cell dynamics is constrained by the condition of homeostasis, clonal tracing studies based on transgenic animal models have advanced our understanding of cell fate behaviour and its dysregulation in disease1,2. But what can be learnt from clonal dynamics in development, where the spatial cohesiveness of clones is impaired by tissue deformations during tissue growth? Drawing on the results of clonal tracing studies, we show that, despite the complexity of organ development, clonal dynamics may converge to a critical state characterized by universal scaling behaviour of clone sizes. By mapping clonal dynamics onto a generalization of the classical theory of aerosols, we elucidate the origin and range of scaling behaviours and show how the identification of universal scaling dependences may allow lineage-specific information to be distilled from experiments. Our study shows the emergence of core concepts of statistical physics in an unexpected context, identifying cellular systems as a laboratory to study non-equilibrium statistical physics.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
Change history
16 March 2018
In the version of this Letter originally published, Steffen Rulands was not listed as a corresponding author. This has been corrected in all versions of the Letter.
References
Kretzschmar, K. & Watt, F. M. Lineage tracing. Cell 148, 33–45 (2012).
Blanpain, C. & Simons, B. D. Unravelling stem cell dynamics by lineage tracing. Nat. Rev. Mol. Cell Biol. 14, 489–502 (2013).
Morrison, S. J. & Spradling, A. C. Stem cells and niches: Mechanisms that promote stem cell maintenance throughout life. Cell 132, 598–611 (2008).
Klein, A. M. & Simons, B. D. Universal patterns of stem cell fate in cycling adult tissues. Development 138, 3103–3111 (2011).
Rulands, S. & Simons, B. D. Tracing cellular dynamics in tissue development, maintenance and disease. Curr. Opin. Cell Biol. 43, 38–45 (2016).
Klein, A. M., Doupé, D. P., Jones, P. H. & Simons, B. D. Mechanism of murine epidermal maintenance: Cell division and the voter model. Phys. Rev. E. 77, 031907 (2008).
Bondue, A. & Blanpain, C. Mesp1: A key regulator of cardiovascular lineage commitment. Circ. Res. 107, 1414–1427 (2010).
Lescroart, F. et al. Early lineage restriction in temporally distinct populations of Mesp1 progenitors during mammalian heart development. Nat. Cell Biol. 16, 829–840 (2014).
Chabab, S. et al. Uncovering the number and clonal dynamics of Mesp1 progenitors during heart morphogenesis. Cell Rep. 14, 1–10 (2016).
Wuidart, A. et al. Quantitative lineage tracing strategies to resolve multipotency in tissue-specific stem cells. Genes Dev. 30, 1261–1277 (2016).
Meilhac, S. M. et al. A retrospective clonal analysis of the myocardium reveals two phases of clonal growth in the developing mouse heart. Development 130, 3877–3889 (2003).
Sedmera, D. & Thompson, R. P. Myocyte proliferation in the developing heart. Dev. Dyn. 240, 1322–1334 (2011).
Foret, L. et al. A general theoretical framework to infer endosomal network dynamics from quantitative image analysis. Curr. Biol. 22, 1381–1390 (2012).
Krapisky, P. L., Redner, S. & Ben-Naim, E. A Kinetic View of Statistical Physics (Cambridge Univ. Press, Cambridge, 2010).
Friedlander, S. K. Smoke, Dust, and Haze (Oxford Univ. Press, Oxford, 2000).
Hendriks, E. M., Ernst, M. H. & Ziff, R. M. Coagulation equations with gelation. J. Stat. Phys. 31, 519–563 (1983).
Ranft, J. et al. Fluidization of tissues by cell division and apoptosis. Proc. Natl Acad. Sci. USA 107, 20863–20868 (2010).
Redner, S. in Disorder and Fracture (NATO ASI Series vol. 204) (eds Charmet, J. C. et al.) (Springer US, Boston, MA, 1990).
Friedlander, S. K. & Wang, C. S. The self-preserving particle size distribution for coagulation by Brownian motion. J. Colloid Interface Sci. 22, 126–132 (1966).
Gupta, V. & Poss, K. D. Clonally dominant cardiomyocytes direct heart morphogenesis. Nature 484, 479–484 (2012).
Clayton, E. et al. A single type of progenitor cell maintains normal epidermis. Nature 446, 185–189 (2007).
Olesen, P., Ferkinghoff-Borg, J., Jensen, M. H. & Mathiesen, J. Diffusion, fragmentation, and coagulation processes: analytical and numerical results. Phys. Rev. E 72, 031103 (2006).
Saga, Y. et al. MesP1 is expressed in the heart precursor cells and required for the formation of a single heart tube. Development 126, 3437–3447 (1999).
Snippert, H. J. et al. Intestinal crypt homeostasis results from neutral competition between symmetrically dividing Lgr5 stem cells. Cell 143, 134–144 (2010).
Schindelin, J. et al. Fiji: an open-source platform for biological-image analysis. Nat. Methods 9, 676–682 (2012).
Acknowledgements
B.D.S. acknowledges the support of the Wellcome Trust (grant number 098357/Z/12/Z). F.L. is supported by a long-term EMBO fellowship and the postdoctoral fellowship of the FNRS. S.C. is supported by a FRIA/FNRS fellowship. C.B. is supported by the ULB, a research grant of the FNRS, the Foundation Bettencourt Schueller, the Foundation ULB and the Foundation Baillet Latour. M.S. is supported by an MRC doctoral training award and A.P. is supported by MRC research grant MR/K018329/1. M.H. is a Wellcome Trust Sir Henry Dale Fellow and is jointly funded by the Wellcome Trust and the Royal Society (104151/Z/14/Z); M.H. and N.P. are funded by a Horizon 2020 grant (LSFM4LIFE). C.H. was funded by a Cambridge Stem Cell Institute Seed funding award for interdisciplinary research awarded to M.H. and B.D.S. We are grateful to K. D. Poss and V. Gupta for making a digital version of their data available to us.
Author information
Authors and Affiliations
Contributions
S.R. and B.D.S. conceived the project. S.C., F.L., C.J.H., N.P. and M.S. performed the experiments and collected the raw data. M.H. supervised the liver experiments. S.R. developed the theory, and performed the modelling and statistical analysis. S.R. and B.D.S drafted the manuscript. All authors edited and approved the final manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethical approval
We have complied with all relevant ethical regulations. Mesp1-Cre mice colonies were maintained in a certified animal facility in accordance with European guidelines. These experiments were approved by the local ethical committee under the reference #LA1230332(CEBEA). Research using mice for pancreas and liver samples has been regulated under the Animal (Scientific Procedures) Act 1986 Amendment Regulations 2012 following ethical review by the University of Cambridge Animal Welfare and Ethical Review Body (AWERB). These experimental data sets were obtained as by-products from other research projects undertaken by the respective laboratories.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary material
Supplementary Figs S1 & S2, Supplementary Theory, Supplementary References 1–10
Rights and permissions
About this article
Cite this article
Rulands, S., Lescroart, F., Chabab, S. et al. Universality of clone dynamics during tissue development. Nature Phys 14, 469–474 (2018). https://doi.org/10.1038/s41567-018-0055-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41567-018-0055-6
This article is cited by
-
A computational model of stem cells’ internal mechanism to recapitulate spatial patterning and maintain the self-organized pattern in the homeostasis state
Scientific Reports (2024)
-
Unconventional colloidal aggregation in chiral bacterial baths
Nature Physics (2023)
-
A computational model of stem cells’ decision-making mechanism to maintain tissue homeostasis and organization in the presence of stochasticity
Scientific Reports (2022)
-
Scale invariance of cell size fluctuations in starving bacteria
Communications Physics (2021)
-
Tracing the cellular basis of islet specification in mouse pancreas
Nature Communications (2020)