Abstract
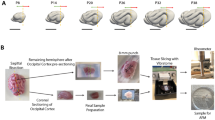
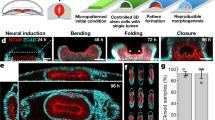
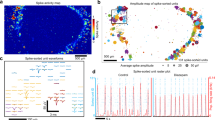
Human brain wrinkling has been implicated in neurodevelopmental disorders and yet its origins remain unknown. Polymer gel models suggest that wrinkling emerges spontaneously due to compression forces arising during differential swelling, but these ideas have not been tested in a living system. Here, we report the appearance of surface wrinkles during the in vitro development and self-organization of human brain organoids in a microfabricated compartment that supports in situ imaging over a timescale of weeks. We observe the emergence of convolutions at a critical cell density and maximal nuclear strain, which are indicative of a mechanical instability. We identify two opposing forces contributing to differential growth: cytoskeletal contraction at the organoid core and cell-cycle-dependent nuclear expansion at the organoid perimeter. The wrinkling wavelength exhibits linear scaling with tissue thickness, consistent with balanced bending and stretching energies. Lissencephalic (smooth brain) organoids display reduced convolutions, modified scaling and a reduced elastic modulus. Although the mechanism here does not include the neuronal migration seen in vivo, it models the physics of the folding brain remarkably well. Our on-chip approach offers a means for studying the emergent properties of organoid development, with implications for the embryonic human brain.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Hannezo, E., Prost, J. & Joanny, J.-F. Mechanical instabilities of biological tubes. Phys. Rev. Lett. 109, 018101 (2012).
Shraiman, B. I. Mechanical feedback as a possible regulator of tissue growth. Proc. Natl. Acad. Sci. USA 102, 3318–3323 (2005).
Klein, Y., Efrati, E. & Sharon, E. Shaping of elastic sheets by prescription of non-Euclidean metrics. Science 315, 1116–1120 (2007).
Savin, T. et al. On the growth and form of the gut. Nature 476, 57–62 (2011).
Kim, H. Y., Varner, V. D. & Nelson, C. M. Apical constriction initiates new bud formation during monopodial branching of the embryonic chicken lung. Development 140, 3146–3155 (2013).
Kiecker, C. & Lumsden, A. Compartments and their boundaries in vertebrate brain development. Nat. Rev. Neurosci. 6, 553–564 (2005).
Taber, L. A. Morphomechanics: transforming tubes into organs. Curr. Opin. Genetics Dev. 27, 7–13 (2014).
Sun, T. & Hevner, R. F. Growth and folding of the mammalian cerebral cortex: from molecules to malformations. Nat. Rev. Neurosci. 15, 217–232 (2014).
Florio, M. & Huttner, W. B. Neural progenitors, neurogenesis and the evolution of the neocortex. Development 141, 2182–2194 (2014).
Mota, B. & Herculano-Houzel, S. Cortical folding scales universally with surface area and thickness, not number of neurons. Science 349, 74–77 (2015).
Reiner, O. et al. Isolation of a Miller–Dieker lissencephaly gene containing G protein beta-subunit-like repeats. Nature 364, 717–721 (1993).
Reiner, O. & Sapir, T. LIS1 functions in normal development and disease. Curr. Opin. Neurobiol. 23, 951–956 (2013).
Groenewold, J. Wrinkling of plates coupled with soft elastic media. Phys. A 298, 32–45 (2001).
Tanaka, T. et al. Mechanical instability of gels at the phase transition. Nature 325, 796–798 (1987).
Bowden, N. et al. Spontaneous formation of ordered structures in thin films of metals supported on an elastomeric polymer. Nature 393, 146–149 (1998).
Cerda, E. & Mahadevan, L. Geometry and physics of wrinkling. Phys. Rev. Lett. 90, 074302 (2003).
Dervaux, J., Couder, Y., Guedeau-Boudeville, M. A. & Ben Amar, M. Shape transition in artificial tumors: From smooth buckles to singular creases. Phys. Rev. Lett. 107, 018103 (2011).
Tallinen, T., Chung, J. Y., Biggins, J. S. & Mahadevan, L. Gyrification from constrained cortical expansion. Proc. Natl Acad. Sci. USA 111, 12667–12672 (2014).
Tallinen, T. et al. On the growth and form of cortical convolutions. Nat. Phys. 12, 588–593 (2016).
Budday, S., Raybaud, C. & Kuhl, E. A mechanical model predicts morphological abnormalities in the developing human brain. Sci. Rep. 4, 5644 (2014).
Gutzman, J. H., Graeden, E. G., Lowery, L. A., Holley, H. S. & Sive, H. Formation of the zebrafish midbrain–hindbrain boundary constriction requires laminin-dependent basal constriction. Mech. Dev. 125, 974–983 (2008).
Lecuit, T. & Lenne, P.-F. Cell surface mechanics and the control of cell shape, tissue patterns and morphogenesis. Nat. Rev. Mol. Cell Biol. 8, 633–644 (2007).
Eiraku, M. et al. Self-organized formation of polarized cortical tissues from ESCs and its active manipulation by extrinsic signals. Cell Stem Cell 3, 519–532 (2008).
Lancaster, M. A. et al. Cerebral organoids model human brain development and microcephaly. Nature 501, 373–379 (2013).
Paşca, A. M. et al. Functional cortical neurons and astrocytes from human pluripotent stem cells in 3D culture. Nat. Methods 12, 671–678 (2015).
Clevers, H. Modeling development and disease with organoids. Cell 165, 1586–1597 (2016).
Bershteyn, M. et al. Human iPSC-derived cerebral organoids model cellular features of lissencephaly and reveal prolonged mitosis of outer radial glia. Cell Stem Cell 20, 435–449.e4 (2017).
Di Lullo, E. & Kriegstein, A. R. The use of brain organoids to investigate neural development and disease. Nat. Rev. Neurosci. 18, 573–584 (2017).
Morel, M., Galas, J.-C., Dahan, M. & Studer, V. Concentration landscape generators for shear free dynamic chemical stimulation. Lab. Chip. 12, 1340–1346 (2012).
Miyata, T., Okamoto, M., Shinoda, T. & Kawaguchi, A. Interkinetic nuclear migration generates and opposes ventricular-zone crowding: insight into tissue mechanics. Front. Cell. Neurosci. 8, 473 (2014).
Hébert, J. M. & Fishell, G. The genetics of early telencephalon patterning: some assembly required. Nat. Rev. Neurosci. 9, 678–685 (2008).
Dantas, T. J., Carabalona, A., Hu, D. J. K. & Vallee, R. B. Emerging roles for motor proteins in progenitor cell behavior and neuronal migration during brain development. Cytoskeleton 73, 566–576 (2016).
Tsai, J.-W., Chen, Y., Kriegstein, A. R. & Vallee, R. B. LIS1 RNA interference blocks neural stem cell division, morphogenesis, and motility at multiple stages. J. Cell Biol. 170, 935–945 (2005).
Schenk, J., Wilsch-Brauninger, M., Calegari, F. & Huttner, W. B. Myosin II is required for interkinetic nuclear migration of neural progenitors. Proc. Natl Acad. Sci. USA 106, 16487–16492 (2009).
Ziv, O. et al. Quantitative live imaging of human embryonic stem cell derived neural rosettes reveals structure–function dynamics coupled to cortical development. PLoS. Comput. Biol. 11, e1004453 (2015).
Bi, W. et al. Increased LIS1 expression affects human and mouse brain development. Nat. Genet. 41, 168–177 (2009).
Reiner, O. LIS1 and DCX: Implications for brain development and human disease in relation to microtubules. Sci. (Cairo) 2013, 393975 (2013).
Sapir, T. Reduction of microtubule catastrophe events by LIS1, platelet-activating factor acetylhydrolase subunit. EMBO J. 16, 6977–6984 (1997).
Faulkner, N. E. A role for the lissencephaly gene LIS1 in mitosis and cytoplasmic dynein function. Nat. Cell Biol. 2, 784–791 (2000).
Feng, Y. et al. LIS1 regulates CNS lamination by interacting with mNudE, a central component of the centrosome. Neuron 28, 665–679 (2000).
Niethammer, M. et al. NUDEL is a novel Cdk5 substrate that associates with LIS1 and cytoplasmic dynein. Neuron 28, 697–711 (2000).
Huang, L., Seker, E., Landers, J. P., Begley, M. R. & Utz, M. Molecular interactions in surface-assembled monolayers of short double-stranded DNA. Langmuir 26, 11574–11580 (2010).
Egan, M. J., Tan, K. & Reck-Peterson, S. L. Lis1 is an initiation factor for dynein-driven organelle transport. J. Cell Biol. 197, 971–982 (2012).
Kardon, J. R. & Vale, R. D. Regulators of the cytoplasmic dynein motor. Nat. Rev. Mol. Cell Biol. 10, 854–865 (2009).
McKenney, R. J., Vershinin, M., Kunwar, A., Vallee, R. B. & Gross, S. P. LIS1 and NudE induce a persistent dynein force-producing state. Cell 141, 304–314 (2010).
Fietz, S. A. et al. Transcriptomes of germinal zones of human and mouse fetal neocortex suggest a role of extracellular matrix in progenitor self-renewal. Proc. Natl Acad. Sci. USA 109, 11836–11841 (2012).
Florio, M. et al. Human-specific gene ARHGAP11B promotes basal progenitor amplification and neocortex expansion. Science 347, 1465–1470 (2015).
Guz, N., Dokukin, M., Kalaparthi, V. & Sokolov, I. If cell mechanics can be described by elastic modulus: Study of different models and probes used in indentation experiments. Biophys. J. 107, 564–575 (2014).
Gholipour, A. et al. A normative spatiotemporal MRI atlas of the fetal brain for automatic segmentation and analysis of early brain growth. Sci. Rep. 7, 476 (2017).
Reillo, I., de Juan Romero, C., García-Cabezas, M. Á. & Borrell, V. A role for intermediate radial glia in the tangential expansion of the mammalian cerebral cortex. Cereb. Cortex. 21, 1674–1694 (2011).
Gafni, O. et al. Derivation of novel human ground state naive pluripotent stem cells. Nature 504, 282–286 (2013).
Cuchiara, M. P., Allen, A. C. B., Chen, T. M., Miller, J. S. & West, J. L. Multilayer microfluidic PEGDA hydrogels. Biomaterials 31, 5491–5497 (2010).
Kim, J.-Y. et al. 3D spherical microtissues and microfluidic technology for multi-tissue experiments and analysis. J. Biotechnol. 205, 24–35 (2015).
Hirotsune, S. et al. Graded reduction of Pafah1b1 (Lis1) activity results in neuronal migration defects and early embryonic lethality. Nat. Genet. 19, 333–339 (1998).
Wu, S. C.-Y. et al. piggyBac is a flexible and highly active transposon as compared to sleeping beauty, Tol2, and Mos1 in mammalian cells. Proc. Natl Acad. Sci. USA 103, 15008–15013 (2006).
Siddiqi, F. et al. Fate mapping by piggyBac transposase reveals that neocortical GLAST+ progenitors generate more astrocytes than Nestin+ progenitors in rat neocortex. Cereb. Cortex. 24, 508–520 (2014).
Shi, Y., Kirwan, P., Smith, J., Robinson, H. P. C. & Livesey, F. J. Human cerebral cortex development from pluripotent stem cells to functional excitatory synapses. Nat. Neurosci. 15, 477–486 (2012). S1.
Jaitin, D. A. et al. Massively parallel single-cell RNA-seq for marker-free decomposition of tissues into cell types. Science 343, 776–779 (2014).
Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet. Journal. 17, 10 (2011).
S., A. FastQC: a quality control tool for high throughput sequence data. Babraham Institute http://www.bioinformatics.babraham.ac.uk/projects/fastqc (2010).
Dobin, A. et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013).
Love, M. I., Huber, W. & Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014).
Ben-Ari Fuchs, S. et al. GeneAnalytics: An integrative gene set analysis tool for next generation sequencing, RNAseq and MicroarrayData. OMICS 20, 139–151 (2016).
Robinson, M. D., McCarthy, D. J. & Smyth, G. K. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140 (2010).
Wu, D. et al. ROAST: rotation gene set tests for complex microarray experiments. Bioinformatics 26, 2176–2182 (2010).
Ma, T. et al. Subcortical origins of human and monkey neocortical interneurons. Nat. Neurosci. 16, 1588–1597 (2013).
Lui, J. H. et al. Radial glia require PDGFD–PDGFRβ signalling in human but not mouse neocortex. Nature 515, 264–268 (2014).
Liu, Y. et al. Medial ganglionic eminence-like cells derived from human embryonic stem cells correct learning and memory deficits. Nat. Biotechnol. 31, 440–447 (2013).
Johnson, M. B. et al. Single-cell analysis reveals transcriptional heterogeneity of neural progenitors in human cortex. Nat. Neurosci. 18, 637–646 (2015).
Close, J. L. et al. Single-cell profiling of an in vitro model of human interneuron development reveals temporal dynamics of cell type production and maturation. Neuron 93, 1035–1048.e5 (2017).
Camp, J. G. et al. Human cerebral organoids recapitulate gene expression programs of fetal neocortex development. Proc. Natl Acad. Sci. USA 112, 15672–15677 (2015).
Lancaster, M. A. et al. Guided self-organization and cortical plate formation in human brain organoids. Nat. Biotechnol. 35, 659–666 (2017).
Onorati, M. et al. Molecular and functional definition of the developing human striatum. Nat. Neurosci. 17, 1804–1815 (2014).
Acknowledgements
We are grateful for the help of S. Viukov, T. Levy and T. Sapir, and for fruitful discussions with S. Safran, A. Tayar and R. Bar-Ziv from the Weizmann Institute of Science. Device fabrication was carried out with assistance from A. Jahanfard, laser microdissection with assistance from Y. Fried, and RNA sequencing and analysis with the advice of H. Keren-Shaul, R. Kohen and T. Olender. Plasmid gifts were received from M. Davidson, Florida State University and J. LoTurco, University of Connecticut. MRI scans were provided by N. Bahi-Buisson, French Institute of Health and Medical Research. O.R. is the incumbent of the Bernstein-Mason Chair of Neurochemistry. E.K. is a Koshland fellow. The research has been supported by the Legacy Heritage Biomedical Program of the Israel Science Foundation (grant no. 2041/16), ERA-NET Neuron with support of the IMOH (grant no. 3-0000-12276), European Cooperation on Science and Technology (COST Action CA16118), Weizmann-FAPESP supported by a research grant from Sergio and Sonia Lozinsky, Nella and Leon Benoziyo Center for Neurological Diseases, Jeanne and Joseph Nissim Foundation for Life Sciences Research, Wohl Biology Endowment Fund, Lulu P. & David J. Levidow Fund for Alzheimers Diseases and Neuroscience Research, the Helen and Martin Kimmel Stem Cell Research Institute, the Kekst Family Institute for Medical Genetics, the David and Fela Shapell Family Center for Genetic Disorders Research. J.H.H. is a New York Stem Cell Foundation (NYSCF)–Robertson Investigator and is supported by research grants from the European Research Council (ERC-CoG2016 CellNaivety), Flight Attendant Medical Research Council (FAMRI), Israel Science Foundation Morasha Program, Nella and Leon Benoziyo Center for Neurological Diseases.
Author information
Authors and Affiliations
Contributions
E.K., A.K. and O.R. planned and conducted experiments. S.C. designed and conducted AFM experiments. J.H. planned and assisted in hES-related experiments. The manuscript was prepared with inputs by all the authors.
Corresponding author
Ethics declarations
Competing interests
J.H.H. is an advisor to Biological Industries Ltd and Accelta Ltd, and had issued patent applications and commercial licences related to certain human stem cell methods used herein.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Movie 1
Developing wild-type organoid. Fluorescence time-lapse images showing nuclear motion and cell division during cell-cycle. Green: Lifeact-GFP, RED: H2B-mCherry. Film duration 36 hours, time steps 10 minutes
Supplementary Movie 2
Developing mutant organoid. Fluorescence time-lapse images showing nuclear motion and cell division in LIS1+/– mutant organoids. Green: Lifeact-GFP, RED: H2B-mCherry. Film duration 5 hours, time steps 3 minutes
Supplementary Information
Supplementary Table S1, Supplementary Movie Captions S1 and S2, Supplementary Figures S1–S21
Rights and permissions
About this article
Cite this article
Karzbrun, E., Kshirsagar, A., Cohen, S.R. et al. Human brain organoids on a chip reveal the physics of folding. Nature Phys 14, 515–522 (2018). https://doi.org/10.1038/s41567-018-0046-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41567-018-0046-7
This article is cited by
-
Neuropathogenesis-on-chips for neurodegenerative diseases
Nature Communications (2024)
-
Genetics of human brain development
Nature Reviews Genetics (2024)
-
Human neuronal maturation comes of age: cellular mechanisms and species differences
Nature Reviews Neuroscience (2024)
-
Humanized brain organoids-on-chip integrated with sensors for screening neuronal activity and neurotoxicity
Microchimica Acta (2024)
-
CRX haploinsufficiency compromises photoreceptor precursor translocation and differentiation in human retinal organoids
Stem Cell Research & Therapy (2023)