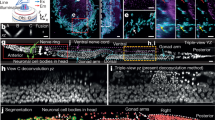
Abstract
Wide-field microscopy of optically thick specimens typically features reduced contrast due to spatial cross-talk, in which the signal at each point in the field of view is the result of a superposition from neighbouring points that are simultaneously illuminated. In 1955, Marvin Minsky proposed confocal microscopy as a solution to this problem. Today, laser scanning confocal fluorescence microscopy is broadly used due to its high depth resolution and sensitivity, but comes at the price of photobleaching, chemical and phototoxicity. Here we present artificial confocal microscopy (ACM) to achieve confocal-level depth sectioning, sensitivity and chemical specificity non-destructively on unlabelled specimens. We equipped a commercial laser scanning confocal instrument with a quantitative phase imaging module, which provides optical path-length maps of the specimen in the same field of view as the fluorescence channel. Using pairs of phase and fluorescence images, we trained a convolution neural network to translate the former into the latter. The training to infer a new tag is very practical as the input and ground truth data are intrinsically registered and the data acquisition is automated. The ACM images present much stronger depth sectioning than the input (phase) images, enabling us to recover confocal-like tomographic volumes of microspheres, hippocampal neurons in culture, and three-dimensional liver cancer spheroids. By training on nucleus-specific tags, ACM allows for segmenting individual nuclei within dense spheroids for both cell counting and volume measurements. In summary, ACM can provide quantitative, dynamic data, non-destructively from thick samples while chemical specificity is recovered computationally.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Data availability
Due to size considerations, the data that support the findings of this study are available from the corresponding author on reasonable request.
Code availability
The code that supports the findings of this study are available from the corresponding author on reasonable request.
References
Freedman, B. S. et al. Modelling kidney disease with CRISPR-mutant kidney organoids derived from human pluripotent epiblast spheroids. Nat. Commun. 6, 1–13 (2015).
Langhans, S. A. Three-dimensional in vitro cell culture models in drug discovery and drug repositioning. Front. Pharmacol. 9, 6 (2018).
Bissell, M. J., Hall, H. G. & Parry, G. How does the extracellular matrix direct gene expression? J. Theor. Biol. 99, 31–68 (1982).
Laschke, M. W. & Menger, M. D. Life is 3D: boosting spheroid function for tissue engineering. Trends Biotechnol. 35, 133–144 (2017).
Bell, C. C. et al. Comparison of hepatic 2D sandwich cultures and 3D spheroids for long-term toxicity applications: a multicenter study. Toxicol. Sci. 162, 655–666 (2018).
Fong, E. L. S., Toh, T. B., Yu, H. & Chow, E. K.-H. 3D culture as a clinically relevant model for personalized medicine. SLAS Tecnol. 22, 245–253 (2017).
Kamm, R. D. et al. Perspective: the promise of multi-cellular engineered living systems. APL Bioeng. 2, 040901 (2018).
Cvetkovic, C. et al. Three-dimensionally printed biological machines powered by skeletal muscle. Proc. Natl Acad. Sci. USA 111, 10125–10130 (2014).
Williams, B. J., Anand, S. V., Rajagopalan, J. & Saif, M. T. A. A self-propelled biohybrid swimmer at low Reynolds number. Nat. Commun. 5, 1–8 (2014).
Chen, X. & Korotkova, O. Optical beam propagation in soft anisotropic biological tissues. Osa Continuum 1, 1055–1067 (2018).
Tuchin, V. V. & Society of Photo-optical Instrumentation Engineers. Tissue Optics: Light Scattering Methods and Instruments for Medical Diagnosis 2nd edn (SPIE/International Society for Optical Engineering, 2007).
Chen, W. et al. High-throughput image analysis of tumor spheroids: a user-friendly software application to measure the size of spheroids automatically and accurately. J Vis Exp. https://doi.org/10.3791/51639 (2014).
Minsky, M. S. Memoir on inventing the confocal. Scanning Microsc. 10, 128–138 (1988).
Wilson, T. & Sheppard, C. Theory and Practice of Scanning Optical Microscopy (Academic, 1984).
Diaspro, A. Optical Fluorescence Microscopy (Springer, 2011).
Lippincott-Schwartz, J., Altan-Bonnet, N. & Patterson, G. H. Photobleaching and photoactivation: following protein dynamics in living cells. Nat. Cell Biol. S7–S14 (2003).
Hoebe, R. A. et al. Controlled light-exposure microscopy reduces photobleaching and phototoxicity in fluorescence live-cell imaging. Nat. Biotechnol. 25, 249–253 (2007).
Crivat, G. & Taraska, J. W. Imaging proteins inside cells with fluorescent tags. Trends Biotechnol. 30, 8–16 (2012).
Graf, B. W. & Boppart, S. A. in Live Cell Imaging: Methods and Protocols 211–227 (Springer, 2010).
North, A. J. Seeing is believing? A beginners’ guide to practical pitfalls in image acquisition. J. Cell Biol. 172, 9–18 (2006).
Hoover, E. E. & Squier, J. A. Advances in multiphoton microscopy technology. Nat. Photon. 7, 93–101 (2013).
Stelzer, E. H. et al. Light sheet fluorescence microscopy. Nat. Rev. Methods Primers 1, 1–25 (2021).
Choi, W. J., Pepple, K. L. & Wang, R. K. Automated three‐dimensional cell counting method for grading uveitis of rodent eye in vivo with optical coherence tomography. J. Biophoton. 11, e201800140 (2018).
Huang, Y. et al. Optical coherence tomography detects necrotic regions and volumetrically quantifies multicellular tumor spheroids. Cancer Res. 77, 6011–6020 (2017).
Schnell, M. et al. High-resolution label-free imaging of tissue morphology with confocal phase microscopy. Optica 7, 1173–1180 (2020).
Hase, E. et al. Scan-less confocal phase imaging based on dual-comb microscopy. Optica 5, 634–643 (2018).
Singh, V. R. et al. Studying nucleic envelope and plasma membrane mechanics of eukaryotic cells using confocal reflectance interferometric microscopy. Nat. Commun. 10, 1–8 (2019).
Liu, C. et al. High-speed line-field confocal holographic microscope for quantitative phase imaging. Opt. Express 24, 9251–9265 (2016).
Popescu, G. Quantitative Phase Imaging of Cells and Tissues (McGraw-Hill, 2011).
Park, Y., Depeursinge, C. & Popescu, G. Quantitative phase imaging in biomedicine. Nat. Photon. 12, 578 (2018).
Chen, X., Kandel, M. E. & Popescu, G. Spatial light interference microscopy: principle and applications to biomedicine. Adv. Opt. Photon. 13, 353–425 (2021).
Chen, X., Kandel, M. E., Hu, C., Lee, Y. J. & Popescu, G. Wolf phase tomography (WPT) of transparent structures using partially coherent illumination. Light Sci. Appl. 9, 1–9 (2020).
Merola, F. et al. Tomographic flow cytometry by digital holography. Light Sci. Appl. 6, e16241 (2017).
Chen, M., Ren, D., Liu, H.-Y., Chowdhury, S. & Waller, L. Multi-layer Born multiple-scattering model for 3D phase microscopy. Optica 7, 394–403 (2020).
Hugonnet, H. et al. Multiscale label-free volumetric holographic histopathology of thick-tissue slides with subcellular resolution. Adv. Photon. 3, 026004 (2021).
Ledwig, P. & Robles, F. E. Epi-mode tomographic quantitative phase imaging in thick scattering samples. Biomed. Opt. Express 10, 3605–3621 (2019).
Kandel, M. E. et al. Epi-illumination gradient light interference microscopy for imaging opaque structures. Nat. Commun. 10, 4691 (2019).
Nguyen, T. H., Kandel, M. E., Rubessa, M., Wheeler, M. B. & Popescu, G. Gradient light interference microscopy for 3D imaging of unlabeled specimens. Nat. Commun. 8, 210 (2017).
Wang, H. et al. Deep learning enables cross-modality super-resolution in fluorescence microscopy. Nat. Methods 16, 103–110 (2019).
Wu, Y. et al. Three-dimensional virtual refocusing of fluorescence microscopy images using deep learning. Nat. Methods 16, 1323–1331 (2019).
Williams, B. M. et al. An artificial intelligence-based deep learning algorithm for the diagnosis of diabetic neuropathy using corneal confocal microscopy: a development and validation study. Diabetologia 63, 419–430 (2020).
Cheng, S., Li, H., Luo, Y., Zheng, Y. & Lai, P. Artificial intelligence-assisted light control and computational imaging through scattering media. J. Innov. Opt. Health Sci. 12, 1930006 (2019).
Ounkomol, C., Seshamani, S., Maleckar, M. M., Collman, F. & Johnson, G. R. Label-free prediction of three-dimensional fluorescence images from transmitted-light microscopy. Nat. Methods 15, 917–920 (2018).
Lee, M. et al. Deep-learning-based three-dimensional label-free tracking and analysis of immunological synapses of CAR-T cells. eLife 9, e49023 (2020).
Tan, M. & Le, Q. EfficientNet: Rethinking Model Scaling for Convolutional Neural Networks. In Proc. 36th International Conference on Machine Learning 6105–6114 (PMLR, 2019).
Chen, X. & Korotkova, O. Probability density functions of instantaneous Stokes parameters on weak scattering. Opt. Commun. 400, 1–8 (2017).
Kanai, Y. & Hirokawa, N. Sorting mechanisms of Tau and MAP2 in neurons: suppressed axonal transit of MAP2 and locally regulated microtubule binding. Neuron 14, 421–432 (1995).
Rein, A. Retroviral RNA packaging: a review. Arch. Viol. Suppl. 9, 513–522 (1994).
Kingma, D.P. & Ba, J. Adam: a method for stochastic optimization. CoRR, abs/1412.6980 (2014).
Deng, J. et al. ImageNet: A large-scale hierarchical image database. In 2009 IEEE Conference on Computer Vision and Pattern Recognition 248–255 (IEEE, 2009).
Kandel, M. E. et al. Multiscale assay of unlabeled neurite dynamics using phase imaging with computational specificity. ACS Sens. 6, 1864–1874 (2021).
Barer, R. Determination of dry mass, thickness, solid and water concentration in living cells. Nature 172, 1097–1098 (1953).
Gonzalez, R. C. & Woods, R. E. Digital Image Processing (Prentice-Hall, 2002).
Stalling, D., Westerhoff M., & Hege H.-C. Amira: A highly interactive system for visual data analysis. Visualization Handb. 38, 749–767 (2005).
Acknowledgements
This work is supported by the National Science Foundation (grant nos. CBET0939511 STC, NRT-UtB 1735252, CBET-1932192), the National Institute of General Medical Sciences (grant no. GM129709), the National Insititute of Neurological Disorders and Stroke (grant nos. NS097610 and NS100019) and the National Cancer Institute (grant no. CA238191).
Author information
Authors and Affiliations
Contributions
X.C., M.E.K., and G.P. conceived the project. X.C. and M.E.K. designed the experiments. X.C. and M.E.K. built the system. X.C. performed imaging. S.H. trained the machine learning network. X.C. and M.E.K. analysed the data. G.T & H.J.C. provided neurons. Y.J.L. cultured neurons and performed immunocytochemistry. K.M.S. & H.K. provided spheroids. X.C., C.H. and G.P. derived the theoretical model. X.C., M.E.K., S.H., C.H. and G.P. wrote the manuscript. M.A. supervised the AI work. G.P. supervised the project.
Corresponding author
Ethics declarations
Competing interests
G.P. had a financial interest in Phi Optics, a company developing QPI technology for materials and life science applications. The remaining authors declare no competing interests.
Peer review
Peer review information
Nature Photonics thanks Adam Wax and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Comparison of ground truth to ACM power spectra from Fig. 3a–l.
Contours circumscribing theoretical resolution limits of confocal fluorescence system (ground truth) are shown in as red dotted circles. The theoretical lateral resolution of the system is 0.22 μm (NA = 1.3, 1 Airy Unit (AU), excitation wavelength at 561 nm), corresponding to a maximum lateral frequency of 14.3 rad/μm. The theoretical axial resolution of the system is about 0.50 μm, corresponding to a maximum axial frequency of 6.3 rad/μm. The 3D frequency coverage of the ground truth and ACM spectra agree, and both reach the theoretical resolution limits.
Supplementary information
Supplementary Information
Supplementary Figs. 1–15 and Notes 1–5.
Supplementary Video 1
ACM-predicted 3D tomography of unlabelled live neurons.
Supplementary Video 2
ACM-predicted timelapse of unlabelled live neurons.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Chen, X., Kandel, M.E., He, S. et al. Artificial confocal microscopy for deep label-free imaging. Nat. Photon. 17, 250–258 (2023). https://doi.org/10.1038/s41566-022-01140-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41566-022-01140-6
This article is cited by
-
On the use of deep learning for phase recovery
Light: Science & Applications (2024)
-
EVATOM: an optical, label-free, machine learning assisted embryo health assessment tool
Communications Biology (2024)
-
Role of Artificial Intelligence in Medical Image Analysis: A Review of Current Trends and Future Directions
Journal of Medical and Biological Engineering (2024)
-
Digital staining in optical microscopy using deep learning - a review
PhotoniX (2023)