Abstract
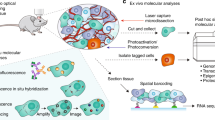
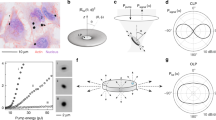
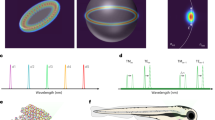
Large-scale single-cell analyses have become increasingly important given the role of cellular heterogeneity in complex biological systems. However, no techniques at present enable optical imaging of uniquely tagged individual cells. Fluorescence-based approaches can distinguish only a small number of distinct cells or cell groups at a time because of spectral crosstalk between conventional fluorophores. Here we investigate large-scale cell tracking using intracellular laser particles as imaging probes that emit coherent laser light with a characteristic wavelength. Made of silica-coated semiconductor microcavities, these laser particles have single-mode emission over a broad range from 1,170 nm to 1,580 nm with sub-nanometre linewidths, enabling massive spectral multiplexing. We explore the stability and biocompatibility of these probes in vitro and their utility for wavelength-multiplexed cell tagging and imaging. We demonstrate real-time tracking of thousands of individual cells in a three-dimensional tumour model over several days, showing different behavioural phenotypes.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
The data that support the plots within this paper and other findings of this study are available from the corresponding author upon reasonable request.
References
Johnson, B. E. et al. Mutational analysis reveals the origin and therapy-driven evolution of recurrent glioma. Science 343, 189–193 (2014).
Wagenblast, E. et al. A model of breast cancer heterogeneity reveals vascular mimicry as a driver of metastasis. Nature 520, 358–362 (2015).
Klein, A. M. M. et al. Droplet barcoding for single-cell transcriptomics applied to embryonic stem cells. Cell 161, 1187–1201 (2015).
Montoro, D. T. et al. A revised airway epithelial hierarchy includes CFTR-expressing ionocytes. Nature 560, 319–324 (2018).
Nguyen, L. V. et al. Barcoding reveals complex clonal dynamics of de novo transformed human mammary cells. Nature 528, 267–271 (2015).
Bhang, H. C. et al. Studying clonal dynamics in response to cancer therapy using high-complexity barcoding. Nat. Med. 21, 440–448 (2015).
Rompolas, P. et al. Live imaging of stem cell and progeny behaviour in physiological hair-follicle regeneration. Nature 487, 496–499 (2012).
Wei, L. et al. Super-multiplex vibrational imaging. Nature 544, 465–470 (2017).
Livet, J. et al. Transgenic strategies for combinatorial expression of fluorescent proteins in the nervous system. Nature 450, 56–62 (2007).
Lin, C. et al. Submicrometre geometrically encoded fluorescent barcodes self-assembled from DNA. Nat. Chem. 4, 832–839 (2012).
Rees, P. et al. Nanoparticle vesicle encoding for imaging and tracking cell populations. Nat. Methods 11, 1177–1181 (2014).
Vahala, K. J. Optical microcavities. Nature 424, 839–846 (2003).
Humar, M. & Yun, S. H. Intracellular microlasers. Nat. Photonics 9, 572–576 (2015).
Schubert, M. et al. Lasing within live cells containing intracellular optical microresonators for barcode-type cell tagging and tracking. Nano Lett. 15, 5647–5652 (2015).
Martino, N. et al. Dense-wavelength-division laser micro-particles: fabrication and imaging in tissues. In Conference on Lasers and Electro-Optics SW3J.4 (Optical Society of America, 2018).
Fikouras, A. H. et al. Non-obstructive intracellular nanolasers. Nat. Commun. 9, 4817 (2018).
Li, E. H. Material parameters of InGaAsP and InAlGaAs systems for use in quantum well structures at low and room temperatures. Phys. E 5, 215–273 (2000).
Smith, A. M., Mancini, M. C. & Nie, S. Bioimaging: second window for in vivo imaging. Nat. Nanotechnol. 4, 710–711 (2009).
Hong, G., Antaris, A. L. & Dai, H. Near-infrared fluorophores for biomedical imaging. Nat. Biomed. Eng. 1, 0010 (2017).
Foreman, M. R., Swaim, J. D. & Vollmer, F. Whispering gallery mode sensors. Adv. Opt. Photonics 7, 168–240 (2015).
Zhang, Z. et al. Visible submicron microdisk lasers. Appl. Phys. Lett. 90, 111119 (2007).
Frateschi, N. C. & Levi, A. F. J. The spectrum of microdisk lasers. J. Appl. Phys. 80, 644–653 (1996).
Andronico, A. et al. Semiconductor microcavities for enhanced nonlinear optics interactions. J. Eur. Opt. Soc. Rapid Publ. 3, 08030 (2008).
Kohl, P. A. Photoelectrochemical etching of semiconductors. IBM J. Res. Dev. 42, 629–638 (1998).
Gil-Santos, E. et al. Scalable high-precision tuning of photonic resonators by resonant cavity-enhanced photoelectrochemical etching. Nat. Commun. 8, 14267 (2017).
Liu, B. S. & Han, M. Synthesis, functionalization, and bioconjugation of monodisperse, silica-coated gold nanoparticles: robust bioprobes. Adv. Funct. Mater. 15, 961–967 (2005).
Choi, W. et al. Tomographic phase microscopy. Nat. Methods 4, 717 (2007).
Intonti, F. et al. Mode tuning of photonic crystal nanocavities by photoinduced non-thermal oxidation. Appl. Phys. Lett. 100, 033116 (2012).
Schöttler, S. et al. Protein adsorption is required for stealth effect of poly(ethylene glycol)- and poly(phosphoester)-coated nanocarriers. Nat. Nanotechnol. 11, 372–377 (2016).
Kim, J., Piao, Y. & Hyeon, T. Multifunctional nanostructured materials for multimodal imaging, and simultaneous imaging and therapy. Chem. Soc. Rev. 38, 372–390 (2009).
Kumar, R. et al. In vivo biodistribution and clearance studies using multimodal organically modified silica nanoparticles. ACS Nano 4, 699–708 (2010).
Patiño, T., Soriano, J., Barrios, L., Ibáñez, E. & Nogués, C. Surface modification of microparticles causes differential uptake responses in normal and tumoral human breast epithelial cells. Sci. Rep. 5, 11371 (2015).
Conner, S. D. & Schmid, S. L. Regulated portals of entry into the cell. Nature 422, 37–44 (2003).
Zhang, T. et al. Cellular effect of high doses of silica-coated quantum dot profiled with high throughput gene expression analysis and high content cellomics measurements. Nano Lett. 6, 800–808 (2006).
Chithrani, B. D. & Chan, W. C. W. Elucidating the mechanism of cellular uptake and removal of protein-coated gold nanoparticles of different sizes and shapes. Nano Lett. 7, 1542–1550 (2007).
Sarkar, D., Ankrum, J. A., Teo, G. S. L., Carman, C. V. & Karp, J. M. Cellular and extracellular programming of cell fate through engineered intracrine-, paracrine-, and endocrine-like mechanisms. Biomaterials 32, 3053–3061 (2011).
Serda, R. E. et al. Mitotic trafficking of silicon microparticles. Nanoscale 1, 250–259 (2009).
Xie, J., Xu, C., Kohler, N., Hou, Y. & Sun, S. Controlled PEGylation of monodisperse Fe3O4 nanoparticles for reduced non‐specific uptake by macrophage cells. Adv. Mater. 19, 3163–3166 (2007).
Cho, S., Humar, M., Martino, N. & Yun, S. H. Laser particle stimulated emission microscopy. Phys. Rev. Lett. 117, 193902 (2016).
Vollmer, F. & Arnold, S. Whispering-gallery-mode biosensing: label-free detection down to single molecules. Nat. Methods 5, 591–596 (2008).
Richards, R., Mason, D., Levy, R., Bearon, R. & See, V. 4D imaging and analysis of multicellular tumour spheroid cell migration and invasion. Preprint at https://www.biorxiv.org/content/10.1101/443648v1 (2018).
Tanay, A. & Regev, A. Scaling single-cell genomics from phenomenology to mechanism. Nature 541, 331–338 (2017).
Wang, X. et al. Three-dimensional intact-tissue sequencing of single-cell transcriptional states. Science 361, eaat5691 (2018).
Kalhor, R. et al. Developmental barcoding of whole mouse via homing CRISPR. Science 361, eaat9804 (2018).
Wong, Y. J. et al. Revisiting the Stober method: inhomogeneity in silica shells. J. Am. Chem. Soc. 133, 11422–11425 (2011).
Palik, E. Optical parameters for the materials in HOC I and HOC II. In Handbook of Optical Constants of Solids Vols 1–5, 313–335 (Elsevier, 1997).
Acknowledgements
This work was supported by the US National Institutes of Health (NIH) grants DP1-OD022296, P41-EB015903, R01-CA192878, and National Science Foundation (NSF) grants ECCS-1505569. Electron microscopy was performed in the Microscopy Core of the Center for Systems Biology/Program in Membrane Biology, which is partially supported by an Inflammatory Bowel Disease Grant DK043351 and a Boston Area Diabetes and Endocrinology Research Center (BADERC) Award DK057521. This research used the resources of the Center for Functional Nanomaterials, which is a US Department of Energy Office of Science User Facility, at Brookhaven National Laboratory under contract number DE-SC0012704 and of the Center for Nanoscale Systems, part of Harvard University, a member of the National Nanotechnology Coordinated Infrastructure Network (NNCI), which is supported by the NSF under award number 1541959. We thank V. Vinarsky and D. Montoro for their comments, and F. Camino for helping with focused ion beam measurements.
Author information
Authors and Affiliations
Contributions
S.-H.Y., N.M. and S.J.J.K. conceived and designed the project. N.M., H.J., H.-M.K., Y.-H.L. and S.-H.Y. designed microdisks. N.M. and A.C.L. fabricated microdisks. N.M. and S.J.J.K. developed the microdisk transfer protocol. S.J.J.K. and S.J.W. developed and performed silica coating of microdisks. N.M. and A.C.L. conducted theoretical simulations. S.J.J.K., S.F. and S.-J.J. performed cell cultures and biocompatibility assays. N.M. and J.W. designed the LASE microscope system. N.M. and S.J.J.K. performed and analysed optical characterization and imaging of LPs. S.J.J.K. and S.-J.J. performed ex vivo lung imaging. S.J.J.K., A.C.L., S.F., S.-J.J. and P.H.D. analysed time-lapse videos of intracellular LPs. N.M., S.J.J.K., A.C.L. and S.-H.Y. prepared figures and wrote the manuscript with input from all authors.
Corresponding author
Ethics declarations
Competing interests
The authors declare the following competing interests: S.-H.Y. holds patents on technologies related to the devices developed in this work. S.-H.Y. and S.J.J.K. have financial interests in LASE Innovation Inc., a company focused on commercializing technologies based on LPs, for a variety of applications in life science and healthcare. S.-H.Y.’s interests were reviewed and are managed by Massachusetts General Hospital and Partners HealthCare in accordance with their conflict of interest policies.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Laser particle optical characterization, biocompatibility, tracking algorithm analysis.
Supplementary Video 1
Uptake of LPs by MDCK-II cells
Supplementary Video 2
Time-lapse brightfield imaging of intracellular LPs
Supplementary Video 3
Time-lapse brightfield imaging of intracellular LPs
Supplementary Video 4
Time-lapse brightfield imaging of intracellular LPs
Supplementary Video 5
Time-lapse imaging of tumour spheroid
Supplementary Video 6
Time-lapse imaging of cell movement in tumour spheroid
Rights and permissions
About this article
Cite this article
Martino, N., Kwok, S.J.J., Liapis, A.C. et al. Wavelength-encoded laser particles for massively multiplexed cell tagging. Nat. Photonics 13, 720–727 (2019). https://doi.org/10.1038/s41566-019-0489-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41566-019-0489-0
This article is cited by
-
Hyperspectral confocal imaging for high-throughput readout and analysis of bio-integrated microlasers
Nature Protocols (2024)
-
High-dimensional multi-pass flow cytometry via spectrally encoded cellular barcoding
Nature Biomedical Engineering (2023)
-
Multipass high-dimensional flow cytometry
Nature Biomedical Engineering (2023)
-
Bridging live-cell imaging and next-generation cancer treatment
Nature Reviews Cancer (2023)
-
A 2D material–based transparent hydrogel with engineerable interference colours
Nature Communications (2022)