Abstract
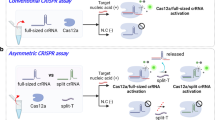
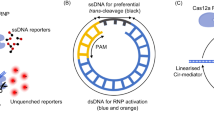
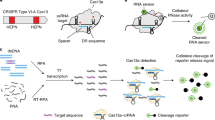
CRISPR-based diagnostics enable specific sensing of DNA and RNA biomarkers associated with human diseases. This is achieved through the binding of guide RNAs to a complementary sequence that activates Cas enzymes to cleave reporter molecules. Currently, most CRISPR-based diagnostics rely on target preamplification to reach sufficient sensitivity for clinical applications. This limits quantification capability and adds complexity to the reaction chemistry. Here we show the combination of a CRISPR–Cas-based reaction with a nanozyme-linked immunosorbent assay, which allows for the quantitative and colorimetric readout of Cas13-mediated RNA detection through catalytic metallic nanoparticles at room temperature (CrisprZyme). We demonstrate that CrisprZyme is easily adaptable to a lateral-flow-based readout and different Cas enzymes and enables the sensing of non-coding RNAs including microRNAs, long non-coding RNAs and circular RNAs. We utilize this platform to identify patients with acute myocardial infarction and to monitor cellular differentiation in vitro and in tissue biopsies from prostate cancer patients. We anticipate that CrisprZyme will serve as a universally applicable signal catalyst for CRISPR-based diagnostics, which will expand the spectrum of targets for preamplification-free, quantitative detection.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Data availability
Research data are available online at https://doi.org/10.5281/zenodo.6553774.
References
Pardee, K. et al. Rapid, low-cost detection of Zika virus using programmable biomolecular components. Cell 165, 1255–1266 (2016).
Gootenberg, J. S. et al. Nucleic acid detection with CRISPR–Cas13a/C2c2. Science 356, 438–442 (2017).
Gootenberg, J. S. et al. Multiplexed and portable nucleic acid detection platform with Cas13, Cas12a, and Csm6. Science 360, 439–444 (2018).
Harrington, L. B. et al. CRISPR–Cas12a target binding unleashes indiscriminate single-stranded DNase activity. Science 360, 436–439 (2018).
Lee, R. A. et al. Ultrasensitive CRISPR-based diagnostic for field-applicable detection of Plasmodium species in symptomatic and asymptomatic malaria. Proc. Natl Acad. Sci. USA 117, 25722–25731 (2020).
Bruch, R. et al. CRISPR/Cas13a-powered electrochemical microfluidic biosensor for nucleic acid amplification-free miRNA diagnostics. Adv. Mater. 31, 1905311 (2019).
Kaminski, M. M. et al. A CRISPR-based assay for the detection of opportunistic infections post-transplantation and for the monitoring of transplant rejection. Nat. Biomed. Eng. 4, 601–609 (2020).
Li, L. et al. HOLMESv2: a CRISPR–Cas12b-assisted platform for nucleic acid detection and DNA methylation quantitation. ACS Synth. Biol. 8, 2228–2237 (2019).
Dai, Y. et al. Exploring the trans-cleavage activity of CRISPR–Cas12a (cpf1) for the development of a universal electrochemical biosensor. Angew. Chem. Int. Ed. 58, 17399–17405 (2019).
Li, S.-Y. et al. CRISPR–Cas12a-assisted nucleic acid detection. Cell Discov. 4, 20 (2018).
Wong, Y.-P., Othman, S., Lau, Y.-L., Radu, S. & Chee, H.-Y. Loop-mediated isothermal amplification (LAMP): a versatile technique for detection of micro-organisms. J. Appl. Microbiol. 124, 626–643 (2018).
Wang, D.-G., Brewster, J. D., Paul, M. & Tomasula, P. M. Two methods for increased specificity and sensitivity in loop-mediated isothermal amplification. Molecules 20, 6048–6059 (2015).
Loynachan, C. N. et al. Platinum nanocatalyst amplification: redefining the gold standard for lateral flow immunoassays with ultrabroad dynamic range. ACS Nano 12, 279–288 (2018).
Kellner, M. J., Koob, J. G., Gootenberg, J. S., Abudayyeh, O. O. & Zhang, F. SHERLOCK: nucleic acid detection with CRISPR nucleases. Nat. Protoc. 14, 2986–3012 (2019).
Kaminski, M. M., Abudayyeh, O. O., Gootenberg, J. S., Zhang, F. & Collins, J. J. CRISPR-based diagnostics. Nat. Biomed. Eng. 5, 643–656 (2021).
Abudayyeh, O. O. et al. RNA targeting with CRISPR–Cas13. Nature 550, 280–284 (2017).
Shan, Y., Zhou, X., Huang, R. & Xing, D. High-fidelity and rapid quantification of miRNA combining crRNA programmability and CRISPR/Cas13a trans-cleavage activity. Anal. Chem. 91, 5278–5285 (2019).
Shi, T., Gao, G. & Cao, Y. Long noncoding RNAs as novel biomarkers have a promising future in cancer diagnostics. Dis. Markers 2016, 9085195 (2016).
Beermann, J., Piccoli, M.-T., Viereck, J. & Thum, T. Non-coding RNAs in development and disease: background, mechanisms, and therapeutic approaches. Physiol. Rev. 96, 1297–1325 (2016).
Wang, C. & Jing, Q. Non-coding RNAs as biomarkers for acute myocardial infarction. Acta Pharmacol. Sin. 39, 1110–1119 (2018).
Zhu, C.-S. et al. Avenues toward microRNA detection in vitro: a review of technical advances and challenges. Comput. Struct. Biotechnol. J. 17, 904–916 (2019).
Dave, V. P. et al. MicroRNA amplification and detection technologies: opportunities and challenges for point of care diagnostics. Lab. Invest. 99, 452–469 (2019).
Garate, X. et al. Identification of the miRNAome of early mesoderm progenitor cells and cardiomyocytes derived from human pluripotent stem cells. Sci. Rep. 8, 8072 (2018).
Chen, S. et al. Widespread and functional RNA circularization in localized prostate cancer. Cell 176, 831–843.e22 (2019).
Burridge, P. W. et al. Chemically defined generation of human cardiomyocytes. Nat. Methods 11, 855–860 (2014).
Acknowledgements
We acknowledge use of the characterization facilities at the Harvey Flower Electron Microscopy Suite (Department of Materials, Imperial College London). We thank the PCBN for providing patient samples. This work was supported by the Department of Defense Prostate Cancer Research Program, US Department of Defense Award Nos W81XWH-18-2-0013, W81XWH-18-2-0015, W81XWH-18-2-0016, W81XWH-18-2-0017, W81XWH-18-2-0018 and W81XWH-18-2-0019 PCRP PCBN. M.B. and M.M.S. acknowledge funding from the British Heart Foundation (grant no. RE/18/4/34215) and the EPSRC IRC in Early Warning Sensing Systems for Infectious Diseases (i-sense) Mobility Fund (grant no. EP/K031953/1). M.M.K. was supported by the German Academy of Sciences, Leopoldina (grant no. LPDS 2018-01) and the Emmy Noether Programme (grant no. KA5060/1-1). M.M.K. is a participant in the BIH Charité Clinician Scientist Program funded by the Charité – Universitätsmedizin Berlin and the Berlin Institute of Health at Charité (BIH). C.A. was supported by the Singapore A*STAR National Science Scholarship fellowship. A.J.S. was supported by the MD Research Stipend of the BIH. H.K. and M.M.S. acknowledge funding from the Research Council of Norway through its Centres of Excellence scheme (grant no. 262613). X.T. is supported by an American Gastroenterological Association Takeda Pharmaceuticals Research Scholar Award in Inflammatory Bowel Disease and the Wyss Institute. J.J.C. was supported by the Paul G. Allen Frontiers Group and the Wyss Institute. M.M.S. acknowledges funding from the EPSRC IRC in Agile Early Warning Sensing Systems for Infectious Diseases and Antimicrobial Resistance (i-sense2) (grant no. EP/R00529X/1), the Royal Academy of Engineering Chair in Emerging Technologies award (no. CiET2021\94) and the Rosetrees Trust. We thank A. Schütz and the team of the Protein Production & Characterization Technology Platform of the Max Delbrück Center for Molecular Medicine in the Helmholtz Association (MDC), Berlin, Germany (https://www.mdc-berlin.de/protein-production-characterization) for producing the LbuCas13a protein.
Author information
Authors and Affiliations
Contributions
M.B., M.M.K., J.J.C. and M.M.S. conceived and designed the research. M.B., M.M.K. and C.A. carried out all the experiments and analysed the data. N.K. performed the LFA experiments, TEM, STEM and EDX imaging and analyses. R.G. and A.J.S. performed RT–RPA, assisted in the design and production of gRNAs and in the optimization of LbuCas13a detection. S.D.-P. optimized and printed LFA strips. X.T. and A.S.D. collected patient samples and edited the manuscript. H.K. performed cell differentiation. M.B., M.M.K., J.J.C. and M.M.S. wrote the manuscript with feedback from all the authors.
Corresponding authors
Ethics declarations
Competing interests
M.M.S., M.B., C.A. and S.D.-P. have filed a patent application (2110729.7) covering the techniques and assay design as described in the manuscript. J.J.C. is a co-founder and director of Sherlock Biosciences. The other authors declare no competing interests.
Peer review
Peer review information
Nature Nanotechnology thanks the anonymous reviewers for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary equation (1), Figs. 1–10 and Tables 1–5.
Rights and permissions
About this article
Cite this article
Broto, M., Kaminski, M.M., Adrianus, C. et al. Nanozyme-catalysed CRISPR assay for preamplification-free detection of non-coding RNAs. Nat. Nanotechnol. 17, 1120–1126 (2022). https://doi.org/10.1038/s41565-022-01179-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41565-022-01179-0
This article is cited by
-
A non-FRET DNA reporter that changes fluorescence colour upon nuclease digestion
Nature Nanotechnology (2024)
-
Topological barrier to Cas12a activation by circular DNA nanostructures facilitates autocatalysis and transforms DNA/RNA sensing
Nature Communications (2024)
-
Biomimetic and bioorthogonal nanozymes for biomedical applications
Nano Convergence (2023)
-
Multiplexed RNA profiling by regenerative catalysis enables blood-based subtyping of brain tumors
Nature Communications (2023)
-
Amplified fluorogenic immunoassay for early diagnosis and monitoring of Alzheimer’s disease from tear fluid
Nature Communications (2023)