Abstract
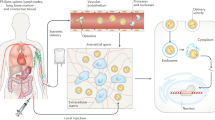
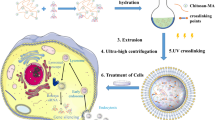
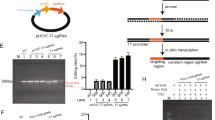
Genome editing holds great potential for cancer treatment due to the ability to precisely inactivate or repair cancer-related genes. However, delivery of CRISPR/Cas to solid tumours for efficient cancer therapy remains challenging. Here we targeted tumour tissue mechanics via a multiplexed dendrimer lipid nanoparticle (LNP) approach involving co-delivery of focal adhesion kinase (FAK) siRNA, Cas9 mRNA and sgRNA (siFAK + CRISPR-LNPs) to enable tumour delivery and enhance gene-editing efficacy. We show that gene editing was enhanced >10-fold in tumour spheroids due to increased cellular uptake and tumour penetration of nanoparticles mediated by FAK-knockdown. siFAK + CRISPR-PD-L1-LNPs reduced extracellular matrix stiffness and efficiently disrupted PD-L1 expression by CRISPR/Cas gene editing, which significantly inhibited tumour growth and metastasis in four mouse models of cancer. Overall, we provide evidence that modulating the stiffness of tumour tissue can enhance gene editing in tumours, which offers a new strategy for synergistic LNPs and other nanoparticle systems to treat cancer using gene editing.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
All data that support the plots within this paper and other findings of this study are shown in the figures and are available from the corresponding author upon reasonable request.
References
Wang, H. X. et al. CRISPR/Cas9-based genome editing for disease modeling and therapy: challenges and opportunities for nonviral delivery. Chem. Rev. 117, 9874–9906 (2017).
Jinek, M. et al. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 337, 816–821 (2012).
Cong, L. et al. Multiplex genome engineering using CRISPR/Cas systems. Science 339, 819–823 (2013).
Mali, P. et al. RNA-guided human genome engineering via Cas9. Science 339, 823–826 (2013).
Xue, W. et al. CRISPR-mediated direct mutation of cancer genes in the mouse liver. Nature 514, 380–384 (2014).
Yin, H. et al. Genome editing with Cas9 in adult mice corrects a disease mutation and phenotype. Nat. Biotechnol. 32, 551–553 (2014).
Maddalo, D. et al. In vivo engineering of oncogenic chromosomal rearrangements with the CRISPR/Cas9 system.Nature 516, 423–427 (2014).
Long, C. Z. et al. Postnatal genome editing partially restores dystrophin expression in a mouse model of muscular dystrophy. Science 351, 400–403 (2016).
Miller, J. B. et al. Non-viral CRISPR/Cas gene editing in vitro and in vivo enabled by synthetic nanoparticle co-delivery of Cas9 mRNA and sgRNA. Angew. Chem. Int. Ed. 56, 1059–1063 (2017).
Wei, T. et al. Delivery of tissue-targeted scalpels: opportunities and challenges for in vivo CRISPR/Cas-based genome editing. ACS Nano 14, 9243–9262 (2020).
Huang, C. H., Lee, K. C. & Doudna, J. A. Applications of CRISPR-Cas enzymes in cancer therapeutics and detection. Trends Cancer 4, 499–512 (2018).
Hanahan, D. & Weinberg, R. A. Hallmarks of cancer: the next generation. Cell 144, 646–674 (2011).
Cox, D. B., Platt, R. J. & Zhang, F. Therapeutic genome editing: prospects and challenges. Nat. Med. 21, 121–131 (2015).
Jiang, H. et al. Targeting focal adhesion kinase renders pancreatic cancers responsive to checkpoint immunotherapy. Nat. Med. 22, 851–860 (2016).
Mohammadi, H. & Sahai, E. Mechanisms and impact of altered tumour mechanics. Nat. Cell Biol. 20, 766–774 (2018).
Humphrey, J. D., Dufresne, E. R. & Schwartz, M. A. Mechanotransduction and extracellular matrix homeostasis. Nat. Rev. Mol. Cell Biol. 15, 802–812 (2014).
Lampi, M. C. & Reinhart-King, C. A. Targeting extracellular matrix stiffness to attenuate disease: from molecular mechanisms to clinical trials. Sci. Trans. Med. 10, eaao0475 (2018).
Seong, J., Wang, N. & Wang, Y. Mechanotransduction at focal adhesions: from physiology to cancer development. J. Cell. Mol. Med. 17, 597–604 (2013).
Serrels, A. et al. Nuclear FAK controls chemokine transcription, Tregs, and evasion of anti-tumor immunity. Cell 163, 160–173 (2015).
Casey, S. C. et al. MYC regulates the antitumor immune response through CD47 and PD-L1. Science 352, 227–231 (2016).
Topalian, S. L., Drake, C. G. & Pardoll, D. M. Targeting the PD-1/B7-H1(PD-L1) pathway to activate anti-tumor immunity. Curr. Opin. Immunol. 24, 207–212 (2012).
Zhou, K. et al. Modular degradable dendrimers enable small RNAs to extend survival in an aggressive liver cancer model. Proc. Natl Acad. Sci. USA 113, 520–525 (2016).
Cheng, Q. et al. Dendrimer-based lipid nanoparticles deliver therapeutic FAH mRNA to normalize liver function and extend survival in a mouse model of hepatorenal tyrosinemia type I. Adv. Mater. 30, e1805308 (2018).
Cheng, Q. et al. Selective ORgan Targeting (SORT) nanoparticles for tissue specific mRNA delivery and CRISPR/Cas gene editing. Nat. Nanotechnol. 15, 313–320 (2020).
Ball, R. L., Hajj, K. A., Vizelman, J., Bajaj, P. & Whitehead, K. A. Lipid nanoparticle formulations for enhanced co-delivery of siRNA and mRNA. Nano Lett. 18, 3814–3822 (2018).
Patel, S. et al. Naturally-occurring cholesterol analogues in lipid nanoparticles induce polymorphic shape and enhance intracellular delivery of mRNA. Nat. Commun. 11, 983 (2020).
Abumanhal-Masarweh, H. et al. Tailoring the lipid composition of nanoparticles modulates their cellular uptake and affects the viability of triple negative breast cancer cells. J. Control. Release 307, 331–341 (2019).
Wei, T., Cheng, Q., Min, Y.-L., Olson, E. N. & Siegwart, D. J. Systemic nanoparticle delivery of CRISPR-Cas9 ribonucleoproteins for effective tissue specific genome editing. Nat. Commun. 11, 3232 (2020).
Liu, S. et al. Membrane-destabilizing ionizable phospholipids for organ-selective mRNA delivery and CRISPR-Cas gene editing. Nat. Mater. 20, 701–710 (2021).
Lee, S. M. et al. A systematic study of unsaturation in lipid nanoparticles leads to improved mRNA transfection in vivo. Angew. Chem. Int. Ed. 60, 5848–5853 (2021).
Mehta, G., Hsiao, A. Y., Ingram, M., Luker, G. D. & Takayama, S. Opportunities and challenges for use of tumor spheroids as models to test drug delivery and efficacy. J. Control. Release 164, 192–204 (2012).
Laklai, H. et al. Genotype tunes pancreatic ductal adenocarcinoma tissue tension to induce matricellular fibrosis and tumor progression. Nat. Med. 22, 497–505 (2016).
Kaksonen, M. & Roux, A. Mechanisms of clathrin-mediated endocytosis. Nat. Rev. Mol. Cell Biol. 19, 313–326 (2018).
Echarri, A. & Del Pozo, M. A. Caveolae–mechanosensitive membrane invaginations linked to actin filaments. J. Cell Sci. 128, 2747–2758 (2015).
Dupont, S. et al. Role of YAP/TAZ in mechanotransduction. Nature 474, 179–183 (2011).
Kraning-Rush, C. M., Califano, J. P. & Reinhart-King, C. A. Cellular traction stresses increase with increasing metastatic potential. PLoS ONE 7, e32572 (2012).
Chaudhuri, O. et al. Extracellular matrix stiffness and composition jointly regulate the induction of malignant phenotypes in mammary epithelium. Nat. Mater. 13, 970–978 (2014).
Hoadley, K. A. et al. Multiplatform analysis of 12 cancer types reveals molecular classification within and across tissues of origin. Cell 158, 929–944 (2014).
Stokes, J. B. et al. Inhibition of focal adhesion kinase by PF-562,271 inhibits the growth and metastasis of pancreatic cancer concomitant with altering the tumor microenvironment. Mol. Cancer Ther. 10, 2135–2145 (2011).
Shachaf, C. M. et al. MYC inactivation uncovers pluripotent differentiation and tumour dormancy in hepatocellular cancer. Nature 431, 1112–1117 (2004).
Zheng, K., Cubero, F. J. & Nevzorova, Y. A. c-MYC making liver sick: role of c-MYC in hepatic cell function, homeostasis and disease. Genes 8, 123 (2017).
Egeblad, M., Rasch, M. G. & Weaver, V. M. Dynamic interplay between the collagen scaffold and tumor evolution. Curr. Opin. Cell Biol. 22, 697–706 (2010).
Paszek, M. J. et al. Tensional homeostasis and the malignant phenotype. Cancer Cell 8, 241–254 (2005).
Fourcade, J. et al. CD8+ T cells specific for tumor antigens can be rendered dysfunctional by the tumor microenvironment through upregulation of the inhibitory receptors BTLA and PD-1. Cancer Res. 72, 887–896 (2012).
Schreiber, R. D., Old, L. J. & Smyth, M. J. Cancer immunoediting: integrating immunity’s roles in cancer suppression and promotion. Science 331, 1565–1570 (2011).
Sica, A. & Mantovani, A. Macrophage plasticity and polarization: in vivo veritas. J. Clin. Invest. 122, 787–795 (2012).
Naito, Y. et al. CD8+ T cells infiltrated within cancer cell nests as a prognostic factor in human colorectal cancer. Cancer Res. 58, 3491–3494 (1998).
Zhang, S. et al. Knockdown of anillin actin binding protein blocks cytokinesis in hepatocytes and reduces liver tumor development in mice without affecting regeneration. Gastroenterology 154, 1421–1434 (2018).
Miller, J. B. & Siegwart, D. J. Design of synthetic materials for intracellular delivery of RNAs: from siRNA-mediated gene silencing to CRISPR/Cas gene editing. Nano Res. 11, 5310–5337 (2018).
Wu, S. Y., Lopez-Berestein, G., Calin, G. A. & Sood, A. K. RNAi therapies: drugging the undruggable. Sci. Trans. Med 6, 240–247 (2014).
Cox, A. D. & Der, C. J. Ras history: the saga continues. Small GTPases 1, 2–27 (2010).
Acknowledgements
D.J.S. acknowledges support from the Cancer Prevention and Research Institute of Texas (CPRIT) (RP190251), the National Institutes of Health (NIH) (R01 EB025192-01A1, R01 CA269787-01), the American Cancer Society (ACS) (RSG-17-012-01) and the Cystic Fibrosis Foundation (CFF) (SIEGWA18XX0). H.Z. acknowledges support from the NIH (R01 DK111588, R01 DK125396, R01 CA251928), the Moody Medical Research Institute and an Emerging Leader Award from the Mark Foundation for Cancer Research (#21-003-ELA). We also acknowledge the UTSW Tissue Resource (National Cancer Institute (5P30CA142543)) and the Moody Foundation Flow Cytometry Facility. T.W. acknowledges a CPRIT Training Grant (RP160157). L.T.J. acknowledges the Pharma Foundation. We are also very grateful to Z. S. Guo (University of Pittsburgh) for sharing ID8-Luc cells with us.
Author information
Authors and Affiliations
Contributions
D.Z. and D.J.S. designed the research. D.Z. designed and performed the experiments. G.W., X.Y., T.W., L.F. and L.T.J., performed the experiments. D.Z., A.M.T., J.X. and Y.H. performed the experiments and data analyses related to mechanical testing. All the authors were involved in the data analyses. D.Z. and D.J.S. wrote the manuscript. H.Z. and all authors discussed and commented on the manuscript.
Corresponding author
Ethics declarations
Competing interests
D.J.S. is a co-founder and consultant of ReCode Therapeutics, which has licensed intellectual property from UT Southwestern. H.Z. has a sponsored research agreement with Alnylam Pharmaceuticals, consults for Flagship Pioneering and serves on the Scientific Advisory Board of Ubiquitix. H.Z.’s interests are not directly related to the contents of this paper. The other authors declare no competing interests.
Peer review
Peer review information
Nature Nanotechnology thanks the anonymous reviewers for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplemental text, methods, Figs. 1–33, Tables 1 and 2, and source data.
Rights and permissions
About this article
Cite this article
Zhang, D., Wang, G., Yu, X. et al. Enhancing CRISPR/Cas gene editing through modulating cellular mechanical properties for cancer therapy. Nat. Nanotechnol. 17, 777–787 (2022). https://doi.org/10.1038/s41565-022-01122-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41565-022-01122-3
This article is cited by
-
Targeting cancer with mRNA–lipid nanoparticles: key considerations and future prospects
Nature Reviews Clinical Oncology (2023)
-
mRNA-based cancer therapeutics
Nature Reviews Cancer (2023)
-
A bimetallic nanoplatform for STING activation and CRISPR/Cas mediated depletion of the methionine transporter in cancer cells restores anti-tumor immune responses
Nature Communications (2023)
-
Lung SORT LNPs enable precise homology-directed repair mediated CRISPR/Cas genome correction in cystic fibrosis models
Nature Communications (2023)
-
Biophysics in tumor growth and progression: from single mechano-sensitive molecules to mechanomedicine
Oncogene (2023)