Abstract
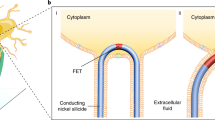
Electrical impulse generation and its conduction within cells or cellular networks are the cornerstone of electrophysiology. However, the advancement of the field is limited by sensing accuracy and the scalability of current recording technologies. Here we describe a scalable platform that enables accurate recording of transmembrane potentials in electrogenic cells. The platform employs a three-dimensional high-performance field-effect transistor array for minimally invasive cellular interfacing that produces faithful recordings, as validated by the gold standard patch clamp. Leveraging the high spatial and temporal resolutions of the field-effect transistors, we measured the intracellular signal conduction velocity of a cardiomyocyte to be 0.182 m s−1, which is about five times the intercellular velocity. We also demonstrate intracellular recordings in cardiac muscle tissue constructs and reveal the signal conduction paths. This platform could provide new capabilities in probing the electrical behaviours of single cells and cellular networks, which carries broad implications for understanding cellular physiology, pathology and cell–cell interactions.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Data availability
All data supporting the findings of this study are available within the paper and its Supplementary Information. The data generated in this study are available from Harvard Dataverse at https://dataverse.harvard.edu/dataset.xhtml?persistentId=doi:10.7910/DVN/7R0DQK&version=DRAFT. Source data are provided with this paper.
Code availability
The code that produced the findings of this study is available from the corresponding author upon reasonable request.
References
Kim, D. H. et al. Dissolvable films of silk fibroin for ultrathin conformal bio-integrated electronics. Nat. Mater. 9, 511–517 (2010).
Abbott, J. et al. A nanoelectrode array for obtaining intracellular recordings from thousands of connected neurons. Nat. Biomed. Eng. 4, 232–241 (2020).
Dai, X. et al. Three-dimensional mapping and regulation of action potential propagation in nanoelectronics-innervated tissues. Nat. Nanotechnol. 11, 776–782 (2016).
Tian, B. et al. Three-dimensional, flexible nanoscale field-effect transistors as localized bioprobes. Science 329, 830–834 (2010).
Jiang, Y. et al. Heterogeneous silicon mesostructures for lipid-supported bioelectric interfaces. Nat. Mater. 15, 1023–1030 (2016).
Wang, X. & Li, M. Automated electrophysiology: high throughput of art. Assay Drug Dev. Technol. 1, 695–708 (2003).
Fast, V. G. & Kléber, A. G. Microscopic conduction in cultured strands of neonatal rat heart cells measured with voltage-sensitive dyes. Circ. Res. 73, 914–925 (1993).
Hong, G. & Lieber, C. M. Novel electrode technologies for neural recordings. Nat. Rev. Neurosci. 20, 330–345 (2019).
Zhang, X. Nanowires pin neurons: a nano "moon landing". Matter 1, 560–562 (2019).
Aranega, A., de la Rosa, A. & Franco, D. Cardiac conduction system anomalies and sudden cardiac death: insights from murine models. Front. Physiol. 3, 211 (2012).
Xu, S. et al. Assembly of micro/nanomaterials into complex, three-dimensional architectures by compressive buckling. Science 347, 154–159 (2015).
Tian, B. & Lieber, C. M. Nanowired bioelectric interfaces. Chem. Rev. 119, 9136–9152 (2019).
Fan, J. A. et al. Fractal design concepts for stretchable electronics. Nat. Commun. 5, 3266 (2014).
Khang, D. Y., Jiang, H., Huang, Y. & Rogers, J. A. A stretchable form of single-crystal silicon for high-performance electronics on rubber substrates. Science 311, 208–212 (2006).
Schaefer, N. et al. Multiplexed neural sensor array of graphene solution-gated field-effect transistors. 2D Mater. 7, 025046 (2020).
Lee, J. W. et al. Analysis of charge sensitivity and low frequency noise limitation in silicon nanowire sensors. J. Appl. Phys. 107, 044501 (2010).
Rettinger, J., Schwarz, S. & Schwarz, W. Electrophysiology (Springer, 2016).
Noy, A. Bionanoelectronics. Adv. Mater. 23, 807–820 (2011).
Hempel, F. et al. PEDOT:PSS organic electrochemical transistor arrays for extracellular electrophysiological sensing of cardiac cells. Biosens. Bioelectron. 93, 132–138 (2017).
Grant, A. O. Cardiac ion channels. Circ. Arrhythm. Electrophysiol. 2, 185–194 (2009).
Duan, X. et al. Intracellular recordings of action potentials by an extracellular nanoscale field-effect transistor. Nat. Nanotechnol. 7, 174–179 (2011).
Gong, H. et al. Biomembrane-modified field effect transistors for sensitive and quantitative detection of biological toxins and pathogens. ACS Nano 13, 3714–3722 (2019).
Qing, Q. et al. Free-standing kinked nanowire transistor probes for targeted intracellular recording in three dimensions. Nat. Nanotechnol. 9, 142–147 (2014).
Zhao, Y. et al. Scalable ultrasmall three-dimensional nanowire transistor probes for intracellular recording. Nat. Nanotechnol. 14, 783–790 (2019).
Abbott, J. et al. CMOS nanoelectrode array for all-electrical intracellular electrophysiological imaging. Nat. Nanotechnol. 12, 460–466 (2017).
Xie, C. et al. Intracellular recording of action potentials by nanopillar electroporation. Nat. Nanotechnol. 7, 185–190 (2012).
Elcarpio, J. O. B. D. et al. HL-1 cells: a cardiac muscle cell line that contracts and retains phenotypic characteristics of the adult cardiomyocyte. Proc. Natl Acad. Sci. USA 95, 2979–2984 (1998).
Hegyi, B., Chen-Izu, Y., Izu, L. T. & Bányász, T. Altered K+ current profiles underlie cardiac action potential shortening in hyperkalemia and β-adrenergic stimulation. Can. J. Physiol. Pharmacol. 97, 773–780 (2019).
Lu, Y.-Y. et al. Electrolyte disturbances differentially regulate sinoatrial node and pulmonary vein electrical activity: a contribution to hypokalemia- or hyponatremia-induced atrial fibrillation. Heart Rhythm 13, 781–788 (2016).
Robinson, J. T. et al. Vertical nanowire electrode arrays as a scalable platform for intracellular interfacing to neuronal circuits. Nat. Nanotechnol. 7, 180–184 (2012).
Czeschik, A. et al. Nanostructured cavity devices for extracellular stimulation of HL-1 cells. Nanoscale 7, 9275–9281 (2015).
Kireev, D. et al. Graphene multielectrode arrays as a versatile tool for extracellular measurements. Adv. Healthc. Mater. 6, 1601433 (2017).
Bers, D. M., Barry, W. H. & Despa, S. Intracellular Na+ regulation in cardiac myocytes. Cardiovasc. Res. 57, 897–912 (2003).
Brown, A. M., Lee, K. S. & Powell, T. Voltage clamp and internal perfusion of single rat heart muscle cells. J. Physiol. 318, 455–477 (1981).
Gouwens, N. W. & Wilson, R. I. Signal propagation in Drosophila central neurons. J. Neurosci. 29, 6239–6249 (2009).
McCain, M. L. et al. Cell-to-cell coupling in engineered pairs of rat ventricular cardiomyocytes: relation between Cx43 immunofluorescence and intercellular electrical conductance. Am. J. Physiol. Heart Circ. Physiol. 302, H443–H450 (2012).
Lancaster, M. A. & Knoblich, J. A. Organogenesis in a dish: modeling development and disease using organoid technologies. Science 345, 1247125 (2014).
Hong, G. et al. A method for single-neuron chronic recording from the retina in awake mice. Science 360, 1447–1451 (2018).
Dipalo, M. et al. Intracellular and extracellular recording of spontaneous action potentials in mammalian neurons and cardiac cells with 3D plasmonic nanoelectrodes. Nano Lett. 17, 3932–3939 (2017).
Nattel, S. Electrical coupling between cardiomyocytes and fibroblasts: experimental testing of a challenging and important concept. Cardiovasc. Res. 114, 349–352 (2018).
Lin, Z. C. et al. Accurate nanoelectrode recording of human pluripotent stem cell-derived cardiomyocytes for assaying drugs and modeling disease. Microsyst. Nanoeng. 3, 16080 (2017).
Desmaisons, D., Vincent, J.-D. & Lledo, P.-M. Control of action potential timing by intrinsic subthreshold oscillations in olfactory bulb output neurons. J. Neurosci. 19, 10727–10737 (1999).
Frohnwieser, B., Chen, L. Q., Schreibmayer, W. & Kallen, R. G. Modulation of the human cardiac sodium channel alpha-subunit by cAMP-dependent protein kinase and the responsible sequence domain. J. Physiol. 498, 309–318 (1997).
Boehmer, G., Greffrath, W., Martin, E. & Hermann, S. Subthreshold oscillation of the membrane potential in magnocellular neurones of the rat supraoptic nucleus. J. Physiol. 526, 115–128 (2000).
Kamiya, K. et al. Electrophysiological measurement of ion channels on plasma/organelle membranes using an on-chip lipid bilayer system. Sci. Rep. 8, 17498 (2018).
Li, J. et al. Scanning microwave microscopy of vital mitochondria in respiration buffer. In Proc. 2018 IEEE MTT-S International Microwave Symposium 115–118 (IEEE, 2018).
Moon, C. H. et al. KR-31378, a novel benzopyran analog, attenuates hypoxia-induced cell death via mitochondrial KATP channel and protein kinase C-ε in heart-derived H9c2 cells. Eur. J. Pharmacol. 506, 27–35 (2004).
Zhao, Y. et al. A platform for generation of chamber-specific cardiac tissues and disease modeling. Cell 176, 913–927.e18 (2019).
Acknowledgements
We thank H. Qian for discussing the cellular electrophysiology measurements taken using the patch clamp, J. Omens for discussing the electrophysiology recordings and electrical stimulation, R. Liu, L. Yan, Y. Luo and H. Ren for discussing the fabrication of the arrayed FETs, Y. Huo for analysing and discussing the characterization of Si doping, M. Makihata for discussing the design of the set-up for measuring the cellular signals, T. Pan for discussing the mechanical simulation, J. Zhao, X. Gao and Y. Wang for discussing data processing and analysis, N. Lawrence for discussing the immunofluorescent staining technique and S. Xiang for preparing the manuscript. This work was supported by the National Institutes of Health (1 R35 GM138250 01).
Author information
Authors and Affiliations
Contributions
S.X., N.K. and Y.G. conceived the idea. S.X., N.K. and Y.G. designed the device. Y.G., N.K., Chunfeng Wang, J.Z., J.M. and Y. Li, fabricated the device. Y.G. and N.K. took the optical and SEM images. Y.G., N.K. and T.M.W. measured the signals with the patch clamp. Y.G., J.S., D.Z., J.M., A.Y. and L.H.-H.H. cultured the HL-1 cells. T.M.W. and J.S. isolated the primary cardiomyocytes of neonatal and adult mice. R.N., S.A. and A.K. prepared the microtissues of neonatal rat cardiomyocytes. J.L. and P.J.B. mapped the FET surface capacitance. Y.G. carried out the finite element analysis simulations. Y.G., N.K., Chunfeng Wang, J.Z. and Z.H. characterized the device electrically. Y.G., N.K., H.G., A.Y. and D.Z. coated the phospholipid on the FET device. Y.G., A.Y. and J.M. performed the fluorescent staining and confocal microscopy imaging. Y.G. fabricated the MEA. Y.G., J.S. and D.Z. carried out the Ca2+ sparks screening assays. Y.G. and D.Z. tested the viability of HL-1 cells. Z.L. and M.L. designed the 10-channel preamplifier. Y.G., Z.H. and W.L. designed the cellular signal measurement set-up. Y.G., Chunfeng Wang, N.K., J.Z. and W.L. measured and analysed the cellular signals. Y.G. and J.Z. drew the schematics and took photographs of the devices. Y.G. carried out the pharmacological and ion-concentration modulation of the HL-1 cells. Y.G. stimulated the HL-1 cells electrically. All authors contributed to discussions on the data and commented on the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information Nature Nanotechnology thanks Xinyan Tracy Cui and Chi Hwan Lee for their contribution to the peer review of this work.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Schematics illustrating the fabrication steps of the 3D FET array.
a, The FET’s drain, source, and gate regions are determined on an n-type SOI wafer by doping. First, the undoped region (that is, the region with only the background doping from the wafer) is coated by a layer of SiO2 as the barrier for spin-on dopants from diffusing into the silicon substrate. Second, the FET shape is defined by photolithography and reactive ion etching. b, The FETs are firstly anchored to the substrate by PTFE, and then are transferred using a PDMS stamp to a temporary 2D substrate coated with PI. c, Multi-layered polymers and metal are coated and patterned on the temporary substrate. Finally, the layered device is transferred to a PDMS stamp and picked up by a water-soluble tape. d, The FET array is transferred to a prestrained elastomer substrate and selectively bonded at the two pre-designed bonding sites. When the prestrain releases, the 2D FET array gets compressed and buckled up to form 3D geometries.
Extended Data Fig. 2 Mapping FET’s conductivity by an atomic force microscope with a bias applied by the scanning tip.
a, A linear scan of the FET’s conductivity was performed with a positive bias applied by the atomic force microscope. The results show a larger positive bias applied on the FET’s lightly doped conduction channel yields a larger conductivity in the channel, which verifies the n-type properties of the FET’s channel, which corresponds to the results by electrical transport characterizations. Scale bar: 2 µm. b, Original data points from the conductivity linear scan showing the FET’s conduction channel turns ‘ON’ at the 1 V tip bias. The ‘threshold voltage’ didn’t appear in the water-gate characterization because we used a much smaller gate bias range for scanning the water-gate characterization. Additionally, we expect the FET device is ‘ON’ at zero bias because the middle region is also n-type with lots of free electrons. The discrepancy between the two measurements is from their different characterization mechanisms, including the gate capacitance, method of applying the gate bias, and the information that can be read from the signals.
Extended Data Fig. 3 Optimizing the FETs’ electrical properties by tuning doping concentrations.
a, Sheet resistances of (i) antimony doped SOI (N-type) and (ii) boron doped SOI (P-type) wafers, determined by high-temperature doping with phosphorus dopant (P509) at 950 °C for various doping times (Δt), which is defined as the period that the apex temperature was held during annealing. b, A typical temperature profile for driving the phosphorus dopants into the SOI wafer. The most effective doping period was at the highest temperature (950 °C), as indicated by Δt. A longer Δt generates a smaller sheet resistance of the SOI. We applied a two-step doping process: the first light doping was for the whole SOI that determined the FET’s conduction channel’s doping concentration; and the second heavy doping was for the whole SOI except the conduction channels. The resultant FET had an N+NN+ structure and worked under a depletion mode. c-e, Transfer characteristics of devices by various doping conditions, including (c) undoped, (d) selectively doped, and (e) heavily uniformly doped sensors. f, Transfer characteristics of (i) p-type and (ii) n-type depletion-mode FETs, showing that the n-type FETs demonstrate about six times larger transconductances (that is, sensitivities) than those p-type FETs. Therefore, we chose n-type depletion-mode FETs in this work.
Extended Data Fig. 4 FETs’ reliability tests under various conditions.
a, i, An FET’s conductance with different pH of the gate solution. The transfer characteristics show high similarities, proving high reliability of the FET under various pH. The common pH for cell culturing is 7.4. ii, Extracted data points at zero gate bias, showing the conductance decreases with increasing the pH, giving another evidence of the n-type conductivity of the FET. b, i, An FET’s transfer characteristics under different temperatures of the gate solution, showing the FET has excellent thermal stability and reliability under various temperatures. The common temperature for cell culturing is 37 °C. ii, Calculated transconductances showing the FET’s transconductance decreases with increasing the temperature, which is due to the effectively reduced mobility of the charge carriers in the conduction channel. c, Parylene C was used as an additional gate dielectric material on top of the SiO2 in the FET, considering SiO2 might be soluble in biological fluids, such as extracellular solutions of cardiac muscle cells. The FETs’ transconductance (that is, sensitivity) barely changed after coating the Parylene C.
Extended Data Fig. 5 Recordings of HL-1 cells’ action potentials by a 10-FET array under electrical stimulation.
a, Changing the stimulation electrode orientation from northwest (nw) to northeast (ne), southwest (sw), and southeast (se) will shift the directions of intercellular signal conductions. The first signal appeared in the FET that is relatively closer to the stimulation electrode and propagated among the cells per their coupling states, as indicated by the black arrows. The intercellular signal conduction velocity under electrical stimulation is from 35.1 to 39.3 μm·ms−1. The variations in the velocity are caused by the fluctuations of temperature, pH value, and ion concentration in the cell culture medium. In all scenarios, the directions of intracellular signal conductions, as indicated by the red arrows, are always the same, that is, from (1,3) to (2,3). b, Schematics showing the latencies of intracellular signal conductions from (1,3) to (2,3).
Extended Data Fig. 6 Small-scale signal conduction within the cardiac tissue by the 128-FET array.
The 128-FET array is classified into 40 units, where each arm has five units. Each unit is labelled by the combination of the located arm name (for example, ‘A’), the loop number (for example, ‘1’), and the relative location (for example, ‘a’). In each unit, the FETs are labelled in i, ii, iii, and iv. Within each unit, intercellular signal conductions via gap junctions in neighboring cells are analyzed, and the latencies are denoted in the heatmaps. The signal conduction velocity inside each unit can be calculated. For instance, in E2b, the signal transmits 70 μm from E2iv to E2vi in 5 ms. By analyzing all signal conductions within each unit, we calculated the small-scale conduction velocity (Supplementary Table 5), whose average and standard deviation are 18.8 ± 7.5 μm·ms−1, which are consistent with previously reported values. The triangle for each unit indicates the selected FET that has the earliest spike within that unit. We use the selected FET in each unit as the reference point to calculate the signal conduction velocities among different units. NaN: Not a number, meaning no cellular signal was recorded.
Supplementary information
Supplementary Information
Materials and Methods, Supplementary Figs. 1–35, Tables 1–5 and references.
41565_2021_1040_MOESM3_ESM.mp4
Supplementary Video 1. From 2D to 3D transformation of the FET array by the compressive buckling technique. We constructed the video by combining a series of optical microscopic images at different stages of the compressive buckling to show the device transformation during compressing and stretching. The transformation is reversible, proving the mechanical robustness of the stretchable device.
41565_2021_1040_MOESM4_ESM.mp4
Supplementary Video 2. Ca2+ sparks of HL-1 cells showing spontaneously firing action potentials without cellular contractions. We carried out such characterizations before using the arrayed FETs to measure the cellular electrical signals. The cells that are spontaneously firing action potentials are pacemakers, similar to the cells in the sinoatrial node of the heart. A pacemaker can transmit the generated action potentials to its electrically coupled cells, triggering these non-pacemaker action potentials. The pacemaker and non-pacemaker cells can be seen as the source and relay of the action potentials.
41565_2021_1040_MOESM5_ESM.mp4
Supplementary Video 3. Quantified Ca2+ sparks of HL-1 cells from Supplementary Video 2 to better illustrate Ca2+ propagative trajectories. Based on image processing and analysis, the video exhibits relatively low spatiotemporal resolution. The Ca2+ sparks appear at the yellow regions. So, by tracking the positions that are yellow, we can identify the signal conduction pathways.
41565_2021_1040_MOESM6_ESM.mp4
Supplementary Video 4. Shift in conduction directions of fluorescent-stained Ca2+ within spontaneously firing HL-1 cells. The conduction directions within the spontaneously firing cells are changing due to temperature, ion concentration or pH fluctuation in the culture medium.
41565_2021_1040_MOESM7_ESM.mp4
Supplementary Video 5. Visualizing signal conductions in each loop. The signals are propagating clockwise, which provides a straightforward validation of the results in Fig. 5e.
Source data
Source Data Fig. 2
Statistical source data.
Source Data Fig. 3
Statistical source data.
Rights and permissions
About this article
Cite this article
Gu, Y., Wang, C., Kim, N. et al. Three-dimensional transistor arrays for intra- and inter-cellular recording. Nat. Nanotechnol. 17, 292–300 (2022). https://doi.org/10.1038/s41565-021-01040-w
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41565-021-01040-w
This article is cited by
-
Graphene-integrated mesh electronics with converged multifunctionality for tracking multimodal excitation-contraction dynamics in cardiac microtissues
Nature Communications (2024)
-
Active Micro-Nano-Collaborative Bioelectronic Device for Advanced Electrophysiological Recording
Nano-Micro Letters (2024)
-
Nature-inspired micropatterns
Nature Reviews Methods Primers (2023)
-
Nanomaterial-based microelectrode arrays for in vitro bidirectional brain–computer interfaces: a review
Microsystems & Nanoengineering (2023)
-
Smartphone-based platforms implementing microfluidic detection with image-based artificial intelligence
Nature Communications (2023)