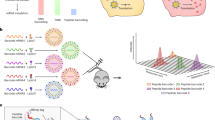
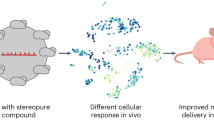
Abstract
Nanoparticles are tested in mice and non-human primates before being selected for clinical trials. Yet the extent to which mRNA delivery, as well as the cellular response to mRNA drug delivery vehicles, is conserved across species in vivo is unknown. Using a species-independent DNA barcoding system, we have compared how 89 lipid nanoparticles deliver mRNA in mice with humanized livers, primatized livers and four controls: mice with ‘murinized’ livers as well as wild-type BL/6, Balb/C and NZB/BlNJ mice. We assessed whether functional delivery results in murine, non-human primate and human hepatocytes can be used to predict delivery in the other species in vivo. By analysing in vivo hepatocytes by RNA sequencing, we identified species-dependent responses to lipid nanoparticles, including mRNA translation and endocytosis. These data support an evidence-based approach to making small-animal preclinical nanoparticle studies more predictive, thereby accelerating the development of RNA therapies.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
All RNA sequencing data have been deposited online at GEO (GSE178313). The scripts used to analyse barcodes are available at Github (https://github.com/Jack-Feldman/barcode_count). All other data are shown in the figures.
References
Adams, D. et al. Patisiran, an RNAi therapeutic, for hereditary transthyretin amyloidosis. N. Engl. J. Med. 379, 11–21 (2018).
Garrelfs, S. F. et al. Lumasiran, an RNAi therapeutic for primary hyperoxaluria type 1. N. Engl. J. Med. 384, 1216–1226 (2021).
Balwani, M. et al. Phase 3 trial of RNAi therapeutic givosiran for acute intermittent porphyria. N. Engl. J. Med. 382, 2289–2301 (2020).
Ray, K. K. et al. Two phase 3 trials of inclisiran in patients with elevated LDL cholesterol. N. Engl. J. Med. 382, 1507–1519 (2020).
Finkel, R. S. et al. Nusinersen versus sham control in infantile-onset spinal muscular atrophy. N. Engl. J. Med. 377, 1723–1732 (2017).
Baden, L. R. et al. Efficacy and safety of the mRNA-1273 SARS-CoV-2 vaccine. N. Engl. J. Med. 384, 403–416 (2020).
Polack, F. P. et al. Safety and efficacy of the BNT162b2 mRNA Covid-19 vaccine. N. Engl. J. Med. 383, 2603–2615 (2020).
Sahin, U. et al. Personalized RNA mutanome vaccines mobilize poly-specific therapeutic immunity against cancer. Nature 547, 222–226 (2017).
Nair, J. K. et al. Multivalent N-acetylgalactosamine-conjugated siRNA localizes in hepatocytes and elicits robust RNAi-mediated gene silencing. J. Am. Chem. Soc. 136, 16958–16961 (2014).
Akinc, A. et al. The Onpattro story and the clinical translation of nanomedicines containing nucleic acid-based drugs. Nat. Nanotechnol. 14, 1084–1087 (2019).
Akinc, A. et al. Targeted delivery of RNAi therapeutics with endogenous and exogenous ligand-based mechanisms. Mol. Ther. 18, 1357–1364 (2010).
Willoughby, J. L. S. et al. Evaluation of GalNAc-siRNA conjugate activity in pre-clinical animal models with reduced asialoglycoprotein receptor expression. Mol. Ther. 26, 105–114 (2018).
Lisowski, L. et al. Selection and evaluation of clinically relevant AAV variants in a xenograft liver model. Nature 506, 382–386 (2014).
Paulk, N. K. et al. Bioengineered AAV capsids with combined high human liver transduction in vivo and unique humoral seroreactivity. Mol. Ther. 26, 289–303 (2018).
Vercauteren, K. et al. Superior in vivo transduction of human hepatocytes using engineered AAV3 capsid. Mol. Ther. 24, 1042–1049 (2016).
Pei, X. et al. Development of AAV variants with human hepatocyte tropism and neutralizing antibody escape capacity. Mol. Ther. Methods Clin. Dev. 18, 259–268 (2020).
Wilson, E. M. et al. Extensive double humanization of both liver and hematopoiesis in FRGN mice. Stem Cell Res. 13, 404–412 (2014).
Foquet, L. et al. Successful engraftment of human hepatocytes in uPA-SCID and FRG® KO mice. Methods Mol. Biol. 1506, 117–130 (2017).
Chen, D. et al. Rapid discovery of potent siRNA-containing lipid nanoparticles enabled by controlled microfluidic formulation. J. Am. Chem. Soc. 134, 6948–6951 (2012).
Sago, C .D. et al. Modifying a commonly expressed endocytic receptor retargets nanoparticles in vivo. Nano Lett. 18, 7590–7600 (2018).
Sago, C. D. et al. Nanoparticles that deliver RNA to bone marrow identified by in vivo directed evolution. J. Am. Chem. Soc. 140, 17095–17105 (2018).
Sago, C. D. et al. High-throughput in vivo screen of functional mRNA delivery identifies nanoparticles for endothelial cell gene editing. Proc. Natl Acad. Sci. USA 115, E9944–E9952 (2018).
Tiwari, P. M. et al. Engineered mRNA-expressed antibodies prevent respiratory syncytial virus infection. Nat. Commun. 9, 3999 (2018).
Paunovska, K. et al. Nanoparticles containing oxidized cholesterol deliver mRNA to the liver microenvironment at clinically relevant doses. Adv. Mater. 31, e1807748 (2019).
Dong, Y. et al. Lipopeptide nanoparticles for potent and selective siRNA delivery in rodents and nonhuman primates. Proc. Natl Acad. Sci. USA 111, 3955–3960 (2014).
Dahlman, J. E. et al. In vivo endothelial siRNA delivery using polymeric nanoparticles with low molecular weight. Nat. Nanotechnol. 9, 648–655 (2014).
Lokugamage, M. P. et al. Mild innate immune activation overrides efficient nanoparticle-mediated RNA delivery. Adv. Mater. 32, 1904905 (2020).
Paunovska, K. et al. Analyzing 2000 in vivo drug delivery data points reveals cholesterol structure impacts nanoparticle delivery. ACS Nano 12, 8341–8349 (2018).
Patel, S. et al. Naturally-occurring cholesterol analogues in lipid nanoparticles induce polymorphic shape and enhance intracellular delivery of mRNA. Nat. Commun. 11, 983 (2020).
Mui, B. L. et al. Influence of polyethylene glycol lipid desorption rates on pharmacokinetics and pharmacodynamics of siRNA lipid nanoparticles. Mol. Ther. Nucleic Acids 2, e139 (2013).
Kaczmarek, J. C. et al. Optimization of a degradable polymer-lipid nanoparticle for potent systemic delivery of mRNA to the lung endothelium and immune cells. Nano Lett. 18, 6449–6454 (2018).
Kranz, L. M. et al. Systemic RNA delivery to dendritic cells exploits antiviral defence for cancer immunotherapy. Nature 534, 396–401 (2016).
Cheng, Q. et al. Selective organ targeting (SORT) nanoparticles for tissue-specific mRNA delivery and CRISPR–Cas gene editing. Nat. Nanotechnol. 15, 313–320 (2020).
Lokugamage, M. P., Sago, C. D. & Dahlman, J. E. Testing thousands of nanoparticles in vivo using DNA barcodes. Curr. Opin. Biomed. Eng. 7, 1–8 (2018).
Patel, S. et al. Boosting intracellular delivery of lipid nanoparticle-encapsulated mRNA. Nano Lett. 17, 5711–5718 (2017).
Picelli, S. et al. Full-length RNA-seq from single cells using Smart-seq2. Nat. Protoc. 9, 171–181 (2014).
Ge, S. X., Son, E. W. & Yao, R. iDEP: an integrated web application for differential expression and pathway analysis of RNA-Seq data. BMC Bioinformatics 19, 534 (2018).
Low, J. Z. B., Khang, T. F. & Tammi, M. T. CORNAS: coverage-dependent RNA-Seq analysis of gene expression data without biological replicates. BMC Bioinformatics 18, 575 (2017).
Huang, D. W., Sherman, B. T. & Lempicki, R. A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4, 44–57 (2009).
Szklarczyk, D. et al. The STRING database in 2017: quality-controlled protein–protein association networks, made broadly accessible. Nucleic Acids Res. 45, D362–D368 (2017).
Dobrovolskaia, M. A., Shurin, M. & Shvedova, A. A. Current understanding of interactions between nanoparticles and the immune system. Toxicol. Appl. Pharmacol. 299, 78–89 (2016).
Azuma, H. et al. Robust expansion of human hepatocytes in Fah–/–/Rag2–/–/Il2rg–/– mice. Nat. Biotechnol. 25, 903–910 (2007).
Acknowledgements
The authors thank K. E. Tiegreen, S. Durham and R. Hughley at Georgia Institute of Technology. The work was funded by the National Institutes of Health (R01-GM132985, awarded to J.E.D., and UG3-TR002855, awarded to J.E.D. and P.J.S.) and DARPA (PREPARE, grant no. HR00111920008, awarded to P.J.S. and J.E.D.).
Author information
Authors and Affiliations
Contributions
M.Z.C.H., C.N.D., P.J.S. and J.E.D. conceived the experiments. P.J.S. and J.E.D. obtained funding for, and oversaw, the research. All authors performed the experiments. M.Z.C.H., C.N.D. and J.E.D. wrote the initial manuscript, which was edited by the authors.
Corresponding author
Ethics declarations
Competing interests
J.E.D. is a consultant for GV. All other authors declare no competing interests.
Peer review
Peer review information
Nature Nanotechnology thanks David Morrissey and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary Figs. 1–10.
Rights and permissions
About this article
Cite this article
Hatit, M.Z.C., Lokugamage, M.P., Dobrowolski, C.N. et al. Species-dependent in vivo mRNA delivery and cellular responses to nanoparticles. Nat. Nanotechnol. 17, 310–318 (2022). https://doi.org/10.1038/s41565-021-01030-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41565-021-01030-y
This article is cited by
-
Strategies to reduce the risks of mRNA drug and vaccine toxicity
Nature Reviews Drug Discovery (2024)
-
Nucleic acid-based drugs for patients with solid tumours
Nature Reviews Clinical Oncology (2024)
-
Nano–Bio Interactions: Exploring the Biological Behavior and the Fate of Lipid-Based Gene Delivery Systems
BioDrugs (2024)
-
A potential paradigm in CRISPR/Cas systems delivery: at the crossroad of microalgal gene editing and algal-mediated nanoparticles
Journal of Nanobiotechnology (2023)
-
Delivery of nucleic acids using nanomaterials
Molecular Biomedicine (2023)