Abstract
Plasma SARS-CoV-2 RNA may represent a viable diagnostic alternative to respiratory RNA levels, which rapidly decline after infection. Quantitative PCR with reverse transcription (RT–qPCR) reference assays exhibit poor performance with plasma, probably reflecting the dilution and degradation of viral RNA released into the circulation, but these issues could be addressed by analysing viral RNA packaged into extracellular vesicles. Here we describe an assay approach in which extracellular vesicles directly captured from plasma are fused with reagent-loaded liposomes to sensitively amplify and detect a SARS-CoV-2 gene target. This approach accurately identified patients with COVID-19, including challenging cases missed by RT–qPCR. SARS-CoV-2-positive extracellular vesicles were detected at day 1 post-infection, and plateaued from day 6 to the day 28 endpoint in a non-human primate model, while signal durations for 20–60 days were observed in young children. This nanotechnology approach uses a non-infectious sample and extends virus detection windows, offering a tool to support COVID-19 diagnosis in patients without SARS-CoV-2 RNA detectable in the respiratory tract.
Similar content being viewed by others
Main
Reverse-transcriptase quantitative polymerase chain reaction (RT–qPCR) analysis of respiratory samples is the gold standard for coronavirus disease 2019 (COVID-19) diagnosis1, but it has limitations. Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2 RNA) levels in the upper respiratory tract rapidly decrease after infection2 while lower respiratory tract levels remain high3. RT–qPCR assays performed after early SARS-CoV-2 infection may thus yield false negatives, but infection events are often unclear, complicating interpretation. Nasopharyngeal tissue highly expresses angiotensin-converting enzyme 2 (ref. 4), the primary receptor for SARS-CoV-2 (ref. 5), but this protein is also expressed in other tissues (for example, cardiac and small intestine6) reported to develop SARS-CoV-2 infections and related pathology7. Nasopharyngeal RT–qPCR assay results thus may not accurately reflect the status of lower respiratory tract or extrapulmonary infections.
Circulating SARS-CoV-2 RNA detectable in mild to severe COVID-19 correlates with and predicts disease severity8,9,10, and appears responsible for extrapulmonary infections7. However, RT–qPCR exhibits poor overall sensitivity for SARS-CoV-2 RNA in plasma (≤41%)11. More sensitive and robust blood-based SARS-CoV-2 RNA assays compatible with routine clinical tests could thus improve COVID-19 diagnosis and prognostic evaluation. However, only two studies employing droplet digital RT–PCR, which is not suited for clinical use, have used ultrasensitive approaches to detect SARS-CoV-2 RNA in serum or plasma.
Infected cells can secrete extracellular vesicles (EVs) containing pathogen-derived factors12,13, protecting these factors from hydrolases and allowing them to accumulate in the circulation14. Hepatitis A and C can infect cells through EV-mediated transfer of their viral genomes15, suggesting that SARS-CoV-2 might use a similar mechanism. Such virus-loaded EVs might thus serve as indicators of systemic viral load and disease severity, but most EV isolation and analysis methods are not feasible for clinical applications.
In this Article, we describe the development of an assay where EVs are directly captured from plasma through the interaction of an antibody with the EV surface protein CD81 and then fused with liposomes containing reagents for reverse transcriptase (RT), recombinase polymerase amplification (RPA) and clustered regularly interspaced short palindromic repeat (CRISPR)–Cas12a reactions using a workflow similar to enzyme-linked immunosorbent assays (ELISAs; Fig. 1a) used for clinical diagnosis. In this assay, guide RNA directs CRISPR–Cas12a binding to an RT–RPA amplicon, inducing concentration-dependent cleavage of a quenched oligonucleotide probe for ultrasensitive detection of SARS-CoV-2 RNA (Extended Data Fig. 1). This approach employs two mature technologies: antibody-mediated capture and liposome-mediated reagent delivery16, with extensive reports in the literature describing liposome synthesis and stabilization methods for clinical applications17.
a, Schematic of the proposed assay, indicating CD81-mediated capture of plasma EVs, their fusion with RT–RPA–CRISPR-loaded liposomes, RT–RPA-mediated target amplification, and signal generation by CRISPR-mediated cleavage of a quenched fluorescent probe in proportion to target amplicon concentration. Analysis sample types include cell culture media and plasma from non-human primate (NHP) COVID-19 disease models and patients with COVID-19. FAM, carboxyfluorescein. b, Schematic of the RT–RPA–CRISPR liposome synthesis workflow and reagents. DMPC, 1,2-dimyristoyl-sn-glycerol-3-phosphorylcholine. c–f, Representative TEM images of liposomes at low (c) and high (d) magnification, and of liposome–EV fusion reactions (e,f). Two repeat experiments were performed. Scale bars, 500 nm (c), 100 nm (d–f). g,h, Schematic (g) and results (h) of an assay measuring the increase in FRET donor signal (588 nm) and decrease in FRET acceptor signal (665 nm) due to dye separation on labelled EVs (2 × 108) as a result of increased distance following membrane fusion after incubation with 1× (2 × 108) or 10× (2 × 109) molar ratios of unlabelled liposomes. The data represent the mean ± s.d. of three replicates. The schematics in a,b and g were created with BioRender.com.
The nanoscale liposomes synthesized to deliver RT–RPA–CRISPR reagents (Fig. 1b) exhibited uniform morphology (~100 nm mean diameter; Extended Data Fig. 2a), and produced fusion products when incubated with purified EVs and poly(ethylene glycol) (PEG8000, ref. 18; Extended Data Fig. 2b,c). Transmission electron microscopy (TEM) analysis of these reactions (Fig. 1c–f) revealed vesicles of ~200 nm, consistent with incomplete fusion (Extended Data Fig. 2c,d). To confirm EV–liposome fusion events, EVs were dual-labelled with 1,1′-dioctadecyl-3,3,3′,3′ tetramethylindocarbocyanine perchlorate (DiI, donor) and 1,1'-dioctadecyl-3,3,3',3'-tetramethylindodicarbocyanine, 4-chlorobenzenesulfonate salt (DiIC18(5); DiD, acceptor) in a Fӧrster resonant energy transfer (FRET) dequenching assay19 (Fig. 1g). FRET activity decreased with the liposome/EV ratio, consistent with the liposome membrane diluting the FRET dyes to respectively enhance and attenuate the donor and acceptor signal (Fig. 1h).
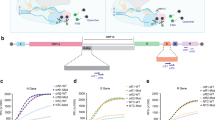
Because EV capture and EV–liposome fusion reactions can occur over broad temperature ranges20,21, we optimized the conditions to produce a one-step CRISPR-enhanced RT–RPA fluorescent detection system (CRISPR-FDS) suitable for incorporation into liposome fusion probes using the assay conditions from a previous study as our starting point22. RT–RPA performed at room temperature (~22 °C) did not induce marked CRISPR-mediated probe conversion under the optimal CRISPR conditions, but a similar and robust CRISPR-FDS signal was observed within the optimal RT–RPA temperature range (Fig. 2a). CRISPR reactions performed at 22, 37 and 42 °C with a constant amount of amplicon revealed different reaction kinetics but similar endpoints, reaching their respective maximum signal intensities at approximately 25, 13 and 10 min (Fig. 2b). RT–RPA–CRISPR-FDS assays performed using these optimized conditions and plasma EV RNA isolates accurately distinguished individuals with and without COVID-19 (Fig. 2c). Negative controls containing genomic RNA from other human respiratory viruses, including two influenza strains and alpha- and betacoronaviruses (Supplementary Table 2) also produced no CRISPR-FDS signal (Fig. 2d), confirming the SARS-CoV-2 specificity of this one-step assay approach.
a,b, CRISPR-FDS assay signals from 5 µl PBS aliquots spiked with 100 copies of SARS-CoV-2 RNA and then incubated at 22, 37 and 42 °C for 15 min with 15 µl RT–RPA reagents and at 37 °C for 15 min with 50 µl CRISPR reagents (a) or at 37 °C for 15 min with 15 µl RT–RPA reagents and then at 22, 37 and 42 °C for 15 min with 50 µl CRISPR reagents (b). c, CRISPR-FDS signals detected with RNA isolated from EVs purified from 50 μl plasma aliquots of individuals diagnosed with or without COVID-19 by positive or negative nasal RT–qPCR results. NC, negative control; PC, positive control. d, CRISPR-FDS signals for RNA extracts obtained from healthy human plasma (50 μl) spiked with or without RNA or virions (>105 copies) of the indicated human respiratory viruses. Human coronavirus (HCoV)-OC43, -HKU1, -229E and -NL63; Middle East respiratory syndrome-associated coronavirus (MERS-CoV); severe acute respiratory syndrome-associated coronavirus (SARS-CoV) and respiratory syncytial virus (RSV). The data represent the mean ± s.d. of three replicates.
Liposomes loaded with RT–RPA–CRISPR reagents were then incubated with antibody-captured EVs to evaluate the CRISPR-FDS signal produced upon vesicle fusion. This analysis found that the CRISPR-FDS signal was notably greater for EVs captured from cells expressing the SARS-CoV-2 N gene instead of control cells (Fig. 3a and Extended Data Fig. 3a,b). In assays analysing EVs directly captured from the plasma of individuals with and without COVID-19, CRISPR-FDS signal differences were also observed 1 h after initiating EV–liposome fusion and increased until the endpoint at 2 h, but did not differ in the absence of fusion (Fig. 3b and Extended Data Fig. 4). Similar results were obtained with a larger patient cohort with similar overall plasma EV levels (Extended Data Fig. 5). Mean CRISPR-FDS intensity was >1.7-fold higher in individuals diagnosed with COVID-19 (Fig. 3c and Supplementary Table 3), but no CRISPR-FDS signal was observed with plasma from patients with other lung diseases (Extended Data Fig. 6). CRISPR-FDS liposome assays also detected EV SARS-CoV-2 RNA in archived plasma from six hospitalized individuals with suspected COVID-19, who had pulmonary CT scans consistent with COVID-19 but negative nasal RT–qPCR results (Fig. 3d). Four of these individuals improved after receiving COVID-19 convalescent plasma (CCP) or had SARS-CoV-2-specific antibodies upon retrospective analysis (Supplementary Data), supporting clinical judgements that all these patients had COVID-19.
a, CRISPR-FDS liposome assay signals detected with EVs isolated from 293F cells stably transfected with a SARS-CoV-2 N gene expression vector or the empty expression vector. The data represent the mean ± s.d. of three replicates. b, CRISPR-FDS liposome assay kinetics for EVs captured from 100 µl plasma aliquots of individuals diagnosed with and without COVID-19 (EVpos and EVneg samples, respectively) upon incubation with CRISPR-FDS reagent-loaded liposomes with or without PEG, or with free CRISPR-FDS reagents. c, CRISPR-FDS liposome assay signals detected from 100 µl plasma aliquots from 20 adults with COVID-19 and 10 adults without COVID-19, as diagnosed by nasal swab RT–qPCR results. d, CRISPR-FDS liposome assay signals detected from plasma samples from six patients diagnosed with COVID-19 but who were diagnosed COVID-19-negative by their nasal swab samples (A40–A45), a healthy donor (HD) and a positive control (whose nasal swab samples were RT–qPCR-positive). Symptoms, imaging data, molecular tests results and treatment responses of patients A40–A45 are summarized in the adjacent table. The data represent the mean ± s.d. of three replicates. CT, computed tomography; NA, not applicable. ****P < 0.0001.
SARS-CoV-2 viral replication is detectable longer in the lower respiratory tract than in the upper respiratory tract23, and other infection sites could give rise to EV SARS-CoV-2 RNA detection in patients with negative nasal RT–qPCR results. To evaluate changes in SARS-CoV-2 RNA levels in nasal tissue and plasma EVs following infection, we analysed serial samples collected from a non-human primate (NHP) model of SARS-CoV-2 infection in which adult African green monkeys were followed for 28 days after exposure to a low-dose SARS-CoV-2 aerosol (Extended Data Fig. 7 and Supplementary Table 5). We observed high nasal SARS-CoV-2 RNA levels by RT–PCR–CRISPR (ref. 24) at day 1 post-infection, which tended to peak between days 1 and 13 post-infection, and decrease rapidly after peak expression (Fig. 4a). CRISPR-FDS liposome assays performed using matched plasma detected lower EV SARS-CoV-2 RNA levels at day 1 post-infection, but these levels were consistently higher at day 6 post-infection and remained stable for the entire time course, suggesting that EV SARS-CoV-2 RNA expression may be a more durable marker of infection.
a, Normalized CRISPR-FDS liposome signal intensity from four NHP (African green monkeys, AG1–4) plasma samples and RT–PCR–CRISPR signal intensity from AG1–AG4 nasal swab samples at the indicated time points. Normalized intensity indicates fold-change versus the baseline mean value. b–d, Positive (red) and negative (blue) results for CRISPR-FDS liposome (EV) fluorescence intensity and results of nasal RT–qPCR and serological (IgG) analyses for three children at the indicated time points after first evaluation. The data represent the mean ± s.d. of three replicates.
Because SARS-CoV-2 RNA levels in NHP plasma EVs exhibited a delayed and sustained peak relative to their nasal swabs, we examined the time course of SARS-CoV-2 EV signals in young children who demonstrated evidence of COVID-19 at or following initial evaluation and who had nasal swab RT–qPCR results and archived blood samples during a >3 month follow-up period. Two children had negative nasal swab results at all time points, but SARS-CoV-2-positive plasma EV results from their initial evaluation to 40–60 days thereafter, with their EV signal remaining positive ~40 days after their first SARS-CoV-2 IgG positive sample24 (Fig. 4c,d). A third child had a single positive RT–qPCR test followed by two negative tests, but four positive plasma EV results over the same period, after which there was a 2 month period in which both tests yielded negative results before again detecting SAR-CoV-2 RNA, suggesting disease recurrence or reinfection (Fig. 4b). However, all plasma samples collected immediately after the positive RT–qPCR results tested positive for EV SARS-CoV-2 RNA. SARS-CoV-2 IgG positive samples were also detected throughout the evaluation period, but this child was only 2 months of age at first evaluation and thus the detected IgG could have derived from the mother, whose infection status was unknown and who did not have samples available for analysis.
Our data indicate that plasma EVs containing SARS-CoV-2 RNA are detectable early after infection and persist after gold-standard nasal RT–qPCR assays return negative results. However, this study does not address several important questions. First, although our data indicate that SARS-CoV-2 RNA is detectable in plasma EVs from early infection onward, the form of this EV RNA cargo is unclear. However, EV-derived SARS-CoV-2 RNA was detected under reaction conditions not expected to disrupt SARS-CoV-2 virions and release their RNA. Proteomic analysis of plasma EVs isolated from patients with COVID-19 also did not detect SARS-CoV-2 viral protein (Extended Data Fig. 8), suggesting that these EVs did not contain substantial amounts of virus-encapsulated SARS-CoV-2 RNA, although the sensitivity of this analysis was limited by the lack of a SARS-CoV-2 RNA-enriched EV sample.
Second, the functional significance of detecting SAR-CoV-2 RNA in these EVs is unclear. Two other single-strand plus-sense RNA viruses are reported to employ the EV biogenesis pathway to deliver their genomes to recipient cells to initiate infection15, but it is not known whether the EVs found to carry SARS-CoV-2 RNA in the current study also support infection. Our CRISPR-FDS liposome assay detects a region of the SARS-CoV-2 N gene, but it is not known whether EVs containing this region also contain the entire intact viral genome required to support infection. Reports indicate that some patients with COVID-19 develop SARS-CoV-2 infections at extrapulmonary sites7, implying that functional virus particles, or possibly EVs that carry the full-length SARS-CoV-2 RNA genome, enter the circulation to initiate infections at secondary sites. SARS-CoV-2 RNA has been detected in the circulation of patients with COVID-19, but the isolation of virus activity from COVID-19 patient plasma or serum has yet to be reported. Given that EVs can directly promote the systemic spread of other viral infections15, similar studies should be conducted for SARS-CoV-2.
Finally, although our results indicate that circulating EVs carry SARS-CoV-2 RNA, our current assay does not detect subgenomic RNA fragments that serve as an indicator of an active infection. Further studies are therefore underway to evaluate SARS-CoV-2 genomic and subgenomic RNA expression in circulating EVs.
Most diagnostics analyse upper respiratory tract samples or saliva, where virus replication appears more transient than in lower respiratory tract, and potentially extrapulmonary, infections25,26. We believe the major utility of our extraction-free approach is its ability to detect plasma EV-derived SARS-CoV-2 RNA as an early and durable sign of systemic infection. EV analysis may be more advantageous than analysis of total plasma RNA for SARS-CoV-2 detection because RNA in captured EVs may contain less degraded or off-target RNA than extravesicular RNA. Our EV assay eliminates the laborious and time-consuming EV isolation steps required by other EV analysis approaches, which produce substantial variation due to batch-to-batch differences in EV yield and purity. Further, it may also be possible to adapt this approach to determine the specific SARS-CoV-2 RNA contribution(s) of potential target tissues during extended infections, which is not possible with analyses that use total plasma RNA.
Some new SARS-CoV-2 diagnostic assays that do not require RNA isolation and/or amplification require high viral loads and analyse upper respiratory tract samples rather than blood. We believe our assay has potential utility as a secondary test for suspected COVID-19 cases that are RT–qPCR-negative but lack alternative diagnoses. It may be particularly valuable for individuals with long-term evidence of infection where transient upper respiratory tract RT–qPCR results may not reflect virus levels in pulmonary or extrapulmonary infections. This includes individuals with compromised immune systems, such as transplant recipients and others receiving immunosuppressive therapies. It may also be relevant during organ donation to reduce the risk of virus transfer, as recently documented for a lung transplant case in which the donor’s SARS-CoV-2 infection was not detected by respiratory tract RT–qPCR testing and led to the death of the recipient and infection of the surgeon27. Our current assay is intended as a clinical application because it analyses plasma, requires wash steps, and uses a benchtop plate reader for its longitudinal readout. However, a portable device that uses a microfluid chip to generate and analyse a fingerstick blood sample could potentially be developed for a point-of-care solution, although this would require stabilization of the reagent-loaded liposomes or an adaptation to analyse the lysates of the captured EVs.
Methods
CRISPR-FDS assay
One tube of TwistAmp Basic powder (TwistDx, TABAS03KIT) was mixed with 35.4 µl rehydration buffer, 3 pmol N gene primer pairs (Supplementary Table 1), 1 µl ProtoScript II reverse transcriptase (NEB M0368L), 3 µl of 80 mM MgOAc and 2.6 µl nuclease-free water. The mixture was aliquoted in three parts for triplicate assays: each 15 µl of RT–RPA reagents was incubated with 5 µl template RNA at 22–42 °C for 15 min and then at 37 °C for 15 min with 50 µl of CRISPR reagents, including 0.16 pmol Cas12a (NEB M0653T), 0.16 pmol N gene guide RNA and 1 pmol FAM-labelled DNA probe for method optimization. The combined RT–RPA–CRISPR assays used for EV RNA detection and pathogen cross activity were performed with a total of 30 min incubation. The nasal swab RT–PCR–CRISPR assays were performed as previously reported24.
CRISPR-FDS liposome probe synthesis
The nanoscale liposomes employed to deliver CRISPR-FDS reagents to EVs were synthesized by dissolving 48 µmol 1,2-dimyristoyl-sn-glycerol-3-phosphorylcholine and 4.8 µmol cholesterol in 1 ml ethanol, which were mixed and dried under nitrogen gas. This material was then mixed with CRISPR-FDS reagents (10 µl RT enzyme, 300 µl 10× NEBuffer 2.1, 0.3 pmol N gene primer pairs, 10 tubes of TwistAmp Basic powder, 300 µl rehydration buffer, 0.16 pmol Cas12a, 0.16 pmol N gene guide RNA and 1 pmol FAM-labelled DNA probe). CRISPR-FDS-loaded liposomes were then prepared by sequentially passing this lipid–reagent mixture through 0.8, 0.4, 0.2 and 0.1 µm polycarbonate membranes (20× for each filter) at room temperature28, after which free reagents and lipid were removed by size-exclusion chromatography using a G25 Dextran column. The liposome fraction was then analysed using a NanoSight NS 300 instrument (Malvern Panalytical) to determine the liposome size distribution, and diluted and vortexed in 5 ml PBS buffer to generate a concentrated liposome solution (8.5 × 109 liposomes per ml), which was aliquoted and stored at 4 °C until the aliquots were diluted for use in CRISPR-FDS liposome assays.
CRISPR-FDS liposome fusion assay
Black-walled 96-well ELISA plates (Corning Costar 3601) coated with 1 µg anti-CD81 (Invitrogen MD5-13548) per well by overnight incubation at room temperature were incubated with 100 µl purified EVs or plasma, as described, for 2 h at 37 °C to allow EV capture, and washed three times with PBS with 0.05% Tween 20. Sample wells were then incubated with 50 µl of a reaction solution containing 4.2 × 108 RT–RPA–CRISPR liposomes, adjusted to a final concentration of 25% mass/volume PEG8000, at 37 °C for 2 h, after which the CRISPR-FDS fluorescent signal was read with a benchtop plate reader (480 nm excitation; 530 nm emission). An EV assay result was considered positive if it was equal to or greater than a cut-off threshold defined by the mean signal of the negative control samples plus three times their standard deviation29.
EV isolation from cell culture medium
EVs were isolated from cell culture medium as previously described30. Briefly, ten 70–80% confluent 15-cm culture dishes of 293F cells (Invitrogen) were washed three times with PBS and then cultured in DMEM medium supplemented with 10% EV-depleted FBS for 48 h, after which the conditioned medium was collected and centrifuged at 2,000g for 30 min to remove cell debris and then passed through a 0.45 μm filter (LG-FPE404150S, LifeGene). The EVs in this clarified supernatant were concentrated by passing this material over a 100 kDa centrifugal filter unit (UFC901008, Thermo Fisher Scientific) at 3,000g for 20–30 min three times. The retained sample was collected by washing the membrane three times with 500 μl PBS and centrifuging twice at 4 °C and 12,000 g for 30 min to precipitate residual debris. This supernatant was then centrifuged twice at 100,000g and 4 °C for 3 h, discarding the supernatant and resuspending the pellet in PBS after each centrifugation step. This EV fraction was then analysed with a NanoSight instrument to determine the EV size distribution, and diluted and vortexed in 5 ml PBS to generate a concentrated EV solution (8.75 × 109 EVs per ml), which was aliquoted and stored at −80 °C until the aliquots were diluted for use in CRISPR-FDS liposome assays.
Plasma EV isolation
The plasma EV samples used in the CRISPR-FDS liposome assays presented in Fig. 3b were isolated with an ExoQuick ULTRA EV Isolation Kit (EQULTRA-20A-1, System Biosciences) following the manufacturer’s instructions. Briefly, 250 µl plasma aliquots were centrifuged at 3,000g for 15 min, the supernatants were then gently mixed with 67 µl ExoQuick solution and incubated on ice for 30 min, and then centrifuged at 3,000g and 4 °C for 10 min. The EV pellets were then processed according to the manufacturer’s instructions, and the EVs were then analysed using a NanoSight instrument.
Particle size measurement
The relative concentrations of the purified plasma EV samples were measured by the bicinchoninic acid assay, and all samples were diluted to a 5 µg ml–1 final concentration in PBS before subsequent analysis. The size distributions and concentrations of the EVs and liposome samples were measured using a NanoSight NS300 instrument employing Nanoparticle Tracking Analysis Software (Malvern Instruments) with a capture duration of 60 s for each sample.
TEM analysis
The liposome or purified EV samples were diluted to a final concentration of ~8.45 × 109 vesicles per μl in 2% phosphotungstic acid (PTA; pH 7.0), to give a final EV concentration of 50 ng EV protein per µl. Sample aliquots (20 µl) were then spotted on parafilm and allowed to adhere for 20 min to a carbon-coated grid that was floated carbon-side down over them, after which excess fluid was removed by wicking with filter paper. The grids were rinsed with distilled water before being placed carbon-side down on a 20 µl drop of filtered 2% PTA (pH 7.0) to stain for approximately 1 min, then the PTA was removed by wicking with filter paper and the samples were allowed to completely dry at room temperature. Images of the liposomes, EVs and vesicle fusions were captured using an FEI TECNAI F30 transmission electron microscope operating at 300 kV.
FRET analysis for liposome–EV fusion
EV aliquots containing 2 × 108 EVs purified from human plasma samples were resuspended in 1 ml PBS containing 5 µl Vybrant DiI (Molecular Probes, V-22885) and 5 µl DiD (Molecular Probes, V-22887), incubated at room temperature for 20 min and then filtered three times through a 100 kDa centrifugal filter unit (UFC901008, Thermo Fisher Scientific) at 3,000g for 20–30 min at room temperature to remove free dyes and concentrate the EVs to a final volume of ~50 µl. Liposome aliquots containing 2 × 108 or 2 × 109 liposomes in 50 µl PBS were mixed with the EVs double-labelled with DiI and DiD, and liposome–EV fusion reactions were performed as described above. The fluorescent signals were excited with a 480 nm laser and fluorescent emission spectra were recorded with a SpectraMax iD5 (Molecular Device) microplate reader from 525 to 750 nm.
SARS-CoV-2 N gene expression vector and cell line
The SARS-CoV-2 N gene was PCR-amplified using a 2019-nCoV_N_Positive Control (IDT 10006625) as the template, and then cloned into the pLenti-CMV-puro lentiviral vector (Addgene 17452) by Gibson assembly (NEB E1602L; Extended Data Fig. 4a). Candidate expression vector subclones expected to carry the N gene target region were validated for full-length sequence identity by Sanger sequencing. Then, 1 × 106 293F cells (Gibco R79007) suspended in 2 ml DMEM with 10% fetal bovine serum were co-transfected with 2 µg pLenti-CMV-puro expression vector with or without the N gene subclone and 1.5 µg psPAX2 (Addgene 12260) and 1 µg pMD2.G (Addgene 12259) vectors. After 48 h, 1 ml of the conditioned culture medium containing lentivirus from the transfected 293F cells was added to the culture wells containing 0.5 × 106 293F cells for 12 h, after which the cells were cultured with 1 µg ml–1 puromycin (Gibco A1113803) for 48 h to select for transduced cells, which were collected and expanded in DMEM with 10% FBS to achieve cell cultures containing 3 × 108 cells for EV isolation, as described above.
Clinical sample collection
The human nasal swab and plasma specimens analysed in this study and demographic data were collected after obtaining prior written informed consent from adult patients or the legal guardians of paediatric patients, who also indicated their assent, in compliance with IRB protocols approved by Tulane University as previously report24. Samples analysed in the adult cohort (Supplementary Table 4) were obtained from patients who had matching plasma and nasal swab samples analysed by the Tulane Molecular Pathology Laboratory between 1 May and 7 May 2020, and whose COVID-19 status was determined on the basis of clinical indications and current CDC guidance. The plasma samples corresponding to the described adult case studies were obtained from individuals who were treated at Tulane Medical Center between 27 April and 14 July 2020 under an institutional review board protocol approved by Tulane University and released to our study team on the same day. Nasal swab results and plasma samples from the indicated child cases were obtained from children who were screened for COVID-19 at the Children’s Hospital New Orleans, Louisiana between March and July 2020 (Supplementary Table 7). The eligibility criteria included any child (≤18 years) receiving care at the Children’s Hospital New Orleans. The clinical results of nasal swabs were determined using the Centers for Disease Control and Prevention 2019-nCoV real-time RT–PCR diagnostic panel.
CCP treatment of adult case studies
Following written informed consent in accordance with the Declaration of Helsinki, ABO-compatible CCP was infused over 1–2 h following pre-medication with 650 mg acetaminophen and 25 mg diphenhydramine. One patient was treated with CCP after obtaining individual emergency investigational new drug (eIND) approval from the US Food and Drug Administration (Fig. 3d patient) were enrolled in the investigator-initiated clinical trial Expanded Access to Convalescent Plasma to Treat and Prevent Pulmonary Complications Associated With COVID-19. This clinical trial was conducted at Tulane University (IND: 020073), approved by the IRB of Tulane University (IRB ref: 2020-595), and registered at clinicaltrials.gov (identifier: NCT04358211).
Plasma and swab collection and processing procedures
Human and NHP blood samples were collected in EDTA tubes and rapidly processed to isolate plasma on the same day. All isolated plasma samples were immediately stored at –80 °C until measurement. All identifying data were removed and samples were assigned a unique subject identification code. Human and NHP nasal swab samples were collected in 200 μl DNA/RNA Shield (R1200, Zymo Research) and stored at −80 °C until processed for RNA. NHP and clinical specimens were processed in an enhanced biosafety level 2/3 space in accordance with a protocol approved by Tulane University Institutional Biosafety Committee. RNA samples were isolated from 100 μl plasma or swab storage buffer using the Zymo Quick-DNA/RNA Viral Kit (D7020) following the assay protocol, and RNA was eluted in 50 μl nuclease-free water and stored at –80 °C until analysis.
COVID-19 IgG test
An ELISA plate was coated with 0.5 µg ml–1 purified SARS-CoV-2 spike protein (kindly provided by Kathryn Hastie at Scripps Research Institute) in 0.1 M NaHCO3 for 1 h at room temperature. The ELISA plate was then washed five times with PBS and EDTA and incubated with blocking buffer (PBS containing 0.5% Tween, 5% dry milk, 4% whey proteins and 10% FBS) for 30 min at 37 °C. In parallel, a set of wells not coated with antigen were incubated with blocking buffer. Plasma was heat-inactivated, diluted 1:100 in blocking buffer and 100 µl per well of diluted plasma was incubated for 1 h at room temperature. The wells were then washed and incubated for 30 min at room temperature with peroxidase-conjugated goat anti-human IgG Fc (Jackson ImmunoResearch 111-035-008) diluted 1:5,000 in blocking buffer, washed with PBS and incubated with 100 µl per well tetramethylbenzidine–H2O2. Colour development was stopped by the addition of 1 M phosphoric acid, and the optical density (OD) was read at 450 nm using a 96-well plate reader. For each sample, the OD values observed from the control wells were subtracted from the OD values observed with S protein to calculate the net OD. Samples with OD > 0.4 were considered positive, based on a cut-off OD value established by preliminary screening of more than 50 pre-COVID19 human sera in which no false positives were detected24.
Animal model
Ethics statement
The Institutional Animal Care and Use Committee of Tulane University reviewed and approved all the procedures for this experiment. The Tulane National Primate Research Center is fully accredited by the Association for Assessment and Accreditation of Laboratory Animal Care. All animals are cared for in accordance with the National Institutes of Health (NIH) guide to Laboratory Animal Care. The Tulane Institutional Biosafety Committee approved the procedures for sample handling, inactivation and removal from biosafety level 3 containment.
Virus information
SARS-CoV-2 isolate USA-WA1/2020 was acquired from BEI Resources, and the collected stock determined to have a 50% tissue culture infective dose (TCID50) per ml of 1 × 106. The virus was passaged in Vero E6 cells in DMEM medium with 2% FBS sequence confirmed by PCR and/or Sanger sequencing. Plaque assays were performed in Vero E6 cells24.
Animals and procedures
Samples from a total of four NHPs aged approximately 7.5 years (four male Chlorocebus aethiops (African green monkeys)) were used for analysis in this study. For viral inoculation, animals were anaesthetized and then acutely exposed by head-only inhalation to SARS-CoV-2 (USA-WA1/2020) resulting in an individual inhaled dose of ~2.5 × 103 TCID50. The animals were biosampled thereafter and observed for 28 days post-infection, including twice-daily monitoring by veterinary staff.
Statistical analysis
CRISPR-FDS and liposome fusion assay signals are expressed as the mean ± s.d. of three or more independent reactions. GraphPad Prism 8 software (GraphPad) was used to calculate one-way analysis of variance (ANOVA) to determine the optimized conditions for the RT–RPA–CRISPR assays. Multiple group comparisons were conducted using one-way ANOVA. Differences were considered statistically significant at P < 0.05.
Reporting Summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Data availability
The data from this work can be obtained from the corresponding authors upon reasonable request. Source data are provided with this paper.
References
Udugama, B. et al. Diagnosing COVID-19: the disease and tools for detection. ACS Nano https://doi.org/10.1021/acsnano.0c02624 (2020).
Liu, Y. et al. Viral dynamics in mild and severe cases of COVID-19. Lancet Infect. Dis. 20, 656–657 (2020).
Wang, W. et al. Detection of SARS-CoV-2 in different types of clinical specimens. JAMA 323, 1843–1844 (2020).
Xu, H. et al. High expression of ACE2 receptor of 2019-nCoV on the epithelial cells of oral mucosa. Int. J. Oral Sci. 12, 8 (2020).
Sungnak, W. et al. SARS-CoV-2 entry factors are highly expressed in nasal epithelial cells together with innate immune genes. Nat. Med. 26, 681–687 (2020).
Burgueño, J. F. et al. Expression of SARS-CoV-2 entry molecules ACE2 and TMPRSS2 in the gut of patients with IBD. Inflamm. Bowel Dis. 26, 797–808 (2020).
Lin, L., Lu, L., Cao, W. & Li, T. Hypothesis for potential pathogenesis of SARS-CoV-2 infection–a review of immune changes in patients with viral pneumonia. Emerg. Microbes Infect. 9, 727–732 (2020).
Hogan, C. A. et al. High frequency of SARS-CoV-2 RNAemia and association with severe disease. Clin. Infect. Dis. https://doi.org/10.1093/cid/ciaa1054 (2020).
Xu, D. et al. Relationship between serum severe acute respiratory syndrome coronavirus 2 nucleic acid and organ damage in coronavirus 2019 patients: a cohort study. Clin. Infect. Dis. https://doi.org/10.1093/cid/ciaa1085 (2020).
Fajnzylber, J. et al. SARS-CoV-2 viral load is associated with increased disease severity and mortality. Nat. Commun. 11, 5493 (2020).
Azghandi, M. & Kerachian, M. A. Detection of novel coronavirus (SARS-CoV-2) RNA in peripheral blood specimens. J. Transl. Med. 18, 412 (2020).
Kuipers, M. E., Hokke, C. H., Smits, H. H. & Nolte-‘t Hoen, E. N. M. Pathogen-derived extracellular vesicle-associated molecules that affect the host immune system: an overview. Front. Microbiol. 9, 2182 (2018).
Rodrigues, M., Fan, J., Lyon, C., Wan, M. & Hu, Y. Role of extracellular vesicles in viral and bacterial infections: pathogenesis, diagnostics, and therapeutics. Theranostics 8, 2709–2721 (2018).
Yáñez-Mó, M. et al. Biological properties of extracellular vesicles and their physiological functions. J. Extracell. Vesicles 4, 27066 (2015).
Longatti, A. The dual role of exosomes in hepatitis A and C virus transmission and viral immune activation. Viruses https://doi.org/10.3390/v7122967 (2015).
Lian, T. & Ho, R. J. Y. Trends and developments in liposome drug delivery systems. J. Pharm. Sci. 90, 667–680 (2001).
Sercombe, L. et al. Advances and challenges of liposome assisted drug delivery. Front. Pharmacol. 6, 286 (2015).
Sato, Y. T. et al. Engineering hybrid exosomes by membrane fusion with liposomes. Sci. Rep. 6, 21933 (2016).
Jumeaux, C. et al. MicroRNA detection by DNA-mediated liposome fusion. ChemBioChem 19, 434–438 (2018).
Gao, X. et al. Rapid detection of exosomal micrornas using virus-mimicking fusogenic vesicles. Angew. Chem. Int. Ed. 58, 8719–8723 (2019).
Lin, Y. et al. Exosome–liposome hybrid nanoparticles deliver CRISPR/Cas9 system in MSCs. Adv. Sci. 5, 1700611 (2018).
Huang, Z. et al. Ultra-sensitive and high-throughput CRISPR-powered COVID-19 diagnosis. Biosens. Bioelectron. 164, 112316 (2020).
Zou, L. et al. SARS-CoV-2 viral load in upper respiratory specimens of infected patients. N. Engl. J. Med. 382, 1177–1179 (2020).
Huang, Z. et al. Sensitive tracking of circulating viral RNA through all stages of SARS-CoV-2 infection. J. Clin. Invest. https://doi.org/10.1172/JCI146031 (2021).
Fozouni, P. et al. Amplification-free detection of SARS-CoV-2 with CRISPR-Cas13a and mobile phone microscopy. Cell 184, 323–333.e9 (2021).
Bruce, E. A. et al. Direct RT-qPCR detection of SARS-CoV-2 RNA from patient nasopharyngeal swabs without an RNA extraction step. PLoS Biol. 18, e3000896 (2020).
Kaul, D. R. et al. Donor to recipient transmission of SARS-CoV-2 by lung transplantation despite negative donor upper respiratory tract testing. Am. J. Transplant. https://doi.org/10.1111/ajt.16532 (2021).
Jin, L., Engelhart, A. E., Adamala, K. P. & Szostak, J. W. Preparation, purification, and use of fatty acid-containing liposomes. J. Vis. Exp. 132, e57324 (2018).
Kim, D., Garner, O. B., Ozcan, A. & Di Carlo, D. Homogeneous entropy-driven amplified detection of biomolecular interactions. ACS Nano 10, 7467–7475 (2016).
Fan, J. et al. Chemoresistance transmission via exosome-mediated EphA2 transfer in pancreatic cancer. Theranostics 8, 5986–5994 (2018).
Acknowledgements
This study was supported by grants R01AI113725, R01AI122932, 1R03AI140977-01 and R21Al126361 from the National Institute of Allergy and Infectious Diseases (NIAID), grant R01HD090927 from the National Institute of Child Health and Human Development, grant 1U01CA252965-01 from the National Cancer Institute and grant W8IXWH1910926 from the Department of Defense. The work at the Tulane National Primate Research Center (TNPRC) was supported in part by NIAID contract no. HHSN272201700033I (C.J.R.) and grant OD011104 from the NIH Office of Research Infrastructure Programs. T.Y.H. also acknowledges the generous support of the Weatherhead Presidential Endowment fund.
Author information
Authors and Affiliations
Contributions
B.N. and T.Y.H. conceived the study, generated the hypotheses and designed the experiments. B.N., Z.H., B.M.Y. and J.F. performed all the experiments and analysed the data. B.N. and C.J.L. wrote the manuscript. J.W.S., K.J.Z., A.N., C.M.B., J.C., X.-M.Y., and N.S.S. conceived and designed the clinical study. C.J.R. conceived and designed the non-human primate study. B.N., C.-z.L., C.J.L. and T.Y.H. performed, analysed and interpreted the clinical data. All authors contributed to the review, revision and finalization of the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information Nature Nanotechnology thanks Arun Richard Chandrasekaran, Graham Cooke and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Detection mechanism and workflow of CRISPR-based RNA detection.
CRISPR-FDS assays utilize simultaneous isothermal reverse transcriptase and RPA reactions to amplify a target region that is then transiently bound in a sequence-specific manner by a CRISPR-Cas12a guide RNA (gRNA) complex, with binding specificity determined by the gRNA sequence specificity (for example, SARS-CoV-2 N gene). Cas12a/gRNA binding activates this enzyme complex to rapidly and nonspecifically cleave an interacting single-stranded polyT DNA oligonucleotide probe present in large molar excess. Cleavage of the assay probe unmasks its quenched fluorescent label to produce fluorescent in proportion to amount of available amplicon in the reaction, which directly reflects the amount of SARS-CoV-2 RNA present in the analysis sample. CRISPR-FDS fluorescent signal development is rapid since probe cleavage occurs in parallel with the amplification of its sequence target, producing a fluorescent signal that can be sensitively read by benchtop plate reader or cellphone-based chip reader., and compared to negative and positive control (NC and PC) samples and concentration standards to detect and quantify the amount of SARS-CoV-2 RNA present in the analyzed sample. CRISPR-FDS assay primers, probes, and gRNAs are listed in Supplementary Table 1.
Extended Data Fig. 2 Characterizations of liposome-EV fusions.
a. Size distribution of RT-RPA-liposomes measured by NanoSight. b. Low-magnification TEM image of plasma-derived EVs used in the liposome-EV fusion reaction showing their size distribution and morphology. Scale bar: 200 nm, two repeat experiments were performed. c. Size distribution of EVs with or without liposome fusion measured by NanoSight. d. High-magnification TEM image of an EV and liposome fusion event indicating multiple vesicle interactions. Scale bar: 200 nm.
Extended Data Fig. 3 SARS-CoV-2 N gene expression in 293 F recombinant cell line.
a. Map of the lentiviral expression vector construct (pLenti-CMV-CoVN-His) that contains the full-length SARS-CoV-2 N gene. b. CRISPR-FDS assay analysis of EV RNA isolated from a 293 F cell line that stably expresses the SARS-CoV-2 N gene from pLenti-CMV-CoVN-His expression vector, which refers to Fig. 3a. Data represent mean ± SD of experimental triplicates. ****, p < 0.0001.
Extended Data Fig. 4 CRISPR-FDS liposome assay kinetics.
CRISPR-FDS liposome assay kinetics detected upon analysis of plasma aliquots (50 µL) from an individual with COVID-19 diagnoses based on positive nasal swab RT-qPCR results. EVs from the subject with positive nasal swab results were at 90 °C for 30 min then incubated with reagent-loaded liposomes (Heat EVs + liposome), CRISPR-FDS reagents not packed into liposomes (Heat EVs + free CRISPR-FDS reagents), EVs without heating were incubated with reagent-loaded liposomes in the absence of PEG (liposome only) or CRISPR-FDS reagents not packed into liposomes (free CRISPR-FDS reagents). Data represent the mean ± SD of three replicates. ****, p < 0.0001.
Extended Data Fig. 5 Relative EV abundance in plasma from patients with COVID-19 (n = 20) and healthy donors (n = 10).
EV ELISA signal obtained from EVs captured from 50 µL plasma with an antibody to the EV surface protein CD81 when captured EVs were sequentially incubated with biotin labelled anti-CD9 antibody, streptavidin-conjugated horseradish peroxidase, and the chromogenic dye tetramethylbenzidine, after which absorbance at 450 nm was measured with a plate-reader. Data are presented as mean ± SD. The box lines indicate the 25, median and 75 percentiles. External lines indicate the minimum and maximum values of each group.
Extended Data Fig. 6 EV COVID-19 assay cross activity evaluation in lung diseases.
Analysis of plasma aliquots (100 µL) from 5 individuals COVID-19 diagnoses based on positive and 5 negative nasal swab RT-qPCR results, 6 pulmonary tuberculosis (PTB) infection 19 pneumonia, 3 Cystic fibrosis (CF), and 1 Allergic Bronchopulmonary Aspergillosis (ABPA). Data are presented as mean ± SD. ****, p < 0.0001. The box lines indicate the 25, median and 75 percentiles. External lines indicate the minimum and maximum values of each group.
Extended Data Fig. 7 NHP model establishment and sample collection.
Timeline for sample collection (plasma and nasal swabs) relative to SARS-CoV-2 infection in the African green monkey COVID-19 model.
Extended Data Fig. 8 Plasma EVs did not demonstrate detectable SARS-CoV-2 protein.
a. Proteomic analysis of plasma exosomes isolated from 3 patients with COVID-19 and 3 healthy donors who were diagnosed by nasal swab RT-qPCR. Numbers indicated the number of EV proteins identified by mass spectrometry from healthy donors and COVID-19 patients, and their overlap when detected peptides were searched against the UniProtKB protein database. No SARS-CoV-2 proteins were identified in this search. b. Western blot analysis of SARS-CoV-2 N protein expression in protein lysates (50 µg/lane) of plasma EVs isolated from three patients with COVID-19 (A1-A3). Positive control (PC), 10 µg SARS-CoV-2 recombinant protein; Negative control (NC), 50 µg EVs isolated from healthy donor plasma.
Supplementary information
Supplementary Information
Supplementary Tables 1–7, Methods and individual case histories for Fig. 3d.
Source data
Source Data Fig. 1
Unprocessed western blots of Extended Data Fig. 8b.
Rights and permissions
About this article
Cite this article
Ning, B., Huang, Z., Youngquist, B.M. et al. Liposome-mediated detection of SARS-CoV-2 RNA-positive extracellular vesicles in plasma. Nat. Nanotechnol. 16, 1039–1044 (2021). https://doi.org/10.1038/s41565-021-00939-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41565-021-00939-8
This article is cited by
-
Thermophoretic glycan profiling of extracellular vesicles for triple-negative breast cancer management
Nature Communications (2024)
-
Exploiting sound for emerging applications of extracellular vesicles
Nano Research (2024)
-
Advanced extracellular vesicle bioinformatic nanomaterials: from enrichment, decoding to clinical diagnostics
Journal of Nanobiotechnology (2023)
-
Genomic communication via circulating extracellular vesicles and long-term health consequences of COVID-19
Journal of Translational Medicine (2023)
-
CRISPR-based assays for low-resource settings
Nature Reviews Bioengineering (2023)