Abstract
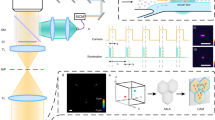
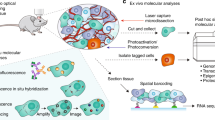
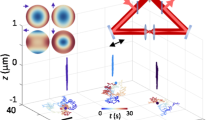
Capturing the dynamics of live cell populations with nanoscale resolution poses a significant challenge, primarily owing to the speed-resolution trade-off of existing microscopy techniques. Flow cytometry would offer sufficient throughput, but lacks subsample detail. Here we show that imaging flow cytometry, in which the point detectors of flow cytometry are replaced with a camera to record 2D images, is compatible with 3D localization microscopy through point-spread-function engineering, which encodes the depth of the emitter into the emission pattern captured by the camera. The extraction of 3D positions from sub-cellular objects of interest is achieved by calibrating the depth-dependent response of the imaging system using fluorescent beads mixed with the sample buffer. This approach enables 4D imaging of up to tens of thousands of objects per minute and can be applied to characterize chromatin dynamics and the uptake and spatial distribution of nanoparticles in live cancer cells.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
Data availability
The data that supports the plots within this paper and other findings of this study are available from the corresponding authors on reasonable request.
Code availability
The analysis scripts for image categorization, calibration, localization and 3D distance measurements were written in MATLAB 2018b (Mathworks) and are available from the corresponding authors on reasonable request.
References
Brown, M. & Wittwer, C. Flow cytometry: principles and clinical applications in hematology. Clin. Chem. 46, 1221–1229 (2000).
Luider, J., Cyfra, M., Johnson, P. & Auer, I. Impact of the New Beckman Coulter Cytomics FC 500 5-Color Flow Cytometer on a regional flow cytometry clinical laboratory service. Lab. Hematol. 10, 102–108 (2004).
Betters, D. M. Use of flow cytometry in clinical practice. J. Adv. Pract. Oncol. 6, 435–440 (2015).
Chandler, W. L., Yeung, W. & Tait, J. F. A new microparticle size calibration standard for use in measuring smaller microparticles using a new flow cytometer. J. Thromb. Haemost. 9, 1216–1224 (2011).
Kay, D. B. & Wheeless, L. L. Laser stroboscopic photography. Technique for cell orientation studies in flow. J. Histochem. Cytochem. 24, 265–268 (1976).
Kay, D. B., Cambier, J. L. & Wheeless, L. L. Imaging in flow. J. Histochem. Cytochem. 27, 329–334 (1979).
Cambier, J. L., Kay, D. B. & Wheeless, L. L. A multidimensional slit-scan flow system. J. Histochem. Cytochem. 27, 321–324 (1979).
George, T. C. et al. Distinguishing modes of cell death using the ImageStream® multispectral imaging flow cytometer. Cytom. Part A. 59A, 237–245 (2004).
Basiji, D. A., Ortyn, W. E., Liang, L., Venkatachalam, V. & Morrissey, P. Cellular image analysis and imaging by flow cytometry. Clin. Lab. Med. 27, 653–670 (2007).
Gorthi, S. S. & Schonbrun, E. Phase imaging flow cytometry using a focus-stack collecting microscope. Opt. Lett. 37, 707 (2012).
Regmi, R., Mohan, K. & Mondal, P. P. High resolution light-sheet based high-throughput imaging cytometry system enables visualization of intra-cellular organelles. AIP Adv. 4, 097125 (2014).
Gualda, E. J., Pereira, H., Martins, G. G., Gardner, R. & Moreno, N. Three-dimensional imaging flow cytometry through light-sheet fluorescence microscopy. Cytom. Part A. 91, 144–151 (2017).
Toprak, E. & Selvin, P. R. New fluorescent tools for watching nanometer-scale conformational changes of single molecules. Annu. Rev. Biophys. Biomol. Struct. 36, 349–369 (2007).
Kao, H. P. & Verkman, A. S. Tracking of single fluorescent particles in three dimensions: use of cylindrical optics to encode particle position. Biophys. J. 67, 1291–1300 (1994).
Shtengel, G. et al. Interferometric fluorescent super-resolution microscopy resolves 3D cellular ultrastructure. Proc. Natl Acad. Sci. USA 106, 3125–3130 (2009).
Juette, M. F. et al. Three-dimensional sub-100 nm resolution fluorescence microscopy of thick samples. Nat. Methods 5, 527–529 (2008).
Abrahamsson, S. et al. Fast multicolor 3D imaging using aberration-corrected multifocus microscopy. Nat. Methods 10, 60–63 (2012).
Backer, A. S. & Moerner, W. E. Extending single-molecule microscopy using optical Fourier processing. J. Phys. Chem. B. 118, 8313–8329 (2014).
Huang, B., Wang, W., Bates, M. & Zhuang, X. Three-dimensional super-resolution imaging by stochastic optical reconstruction microscopy. Science 319, 810–813 (2008).
Mlodzianoski, M. J., Juette, M. F., Beane, G. L. & Bewersdorf, J. Experimental characterization of 3D localization techniques for particle-tracking and super-resolution microscopy. Opt. Express 17, 8264 (2009).
Dowski, E. R. & Cathey, W. T. Extended depth of field through wave-front coding. Appl. Opt. 34, 1859 (1995).
Ortyn, W. E. et al. Extended depth of field imaging for high speed cell analysis. Cytometry. A. 71, 215–231 (2007).
Shechtman, Y., Weiss, L. E., Backer, A. S., Sahl, S. J. & Moerner, W. E. Precise three-dimensional scan-free multiple-particle tracking overlarge axial ranges with tetrapod point spread functions. Nano Lett. 15, 4194–4199 (2015).
Cierpka, C., Rossi, M., Segura, R. & Kähler, C. J. On the calibration of astigmatism particle tracking velocimetry for microflows. Meas. Sci. Technol. 22, 015401 (2011).
Schmied, J. J. et al. DNA origami–based standards for quantitative fluorescence microscopy. Nat. Protoc. 9, 1367 (2014).
Finn, E. H. et al. Extensive heterogeneity and intrinsic variation in spatial genome organization. Cell 176, 1502–1515.e10 (2019).
Aylon, Y. & Kupiec, M. New insights into the mechanism of homologous recombination in yeast. Mutat. Res. 566, 231–248 (2004).
Dultz, E. et al. Quantitative imaging of chromatin decompaction in living cells. Mol. Biol. Cell 29, 1763–1777 (2018).
Khanna, N., Zhang, Y., Lucas, J. S., Dudko, O. K. & Murre, C. Chromosome dynamics near the sol-gel phase transition dictate the timing of remote genomic interactions. Nat. Commun. 10, 2771 (2019).
Even-Faitelson, L., Hassan-Zadeh, V., Baghestani, Z. & Bazett-Jones, D. P. Coming to terms with chromatin structure. Chromosoma 125, 95–110 (2016).
Hopper, J. E., Broach, J. R. & Rowe, L. B. Regulation of the galactose pathway in Saccharomyces cerevisiae: induction of uridyl transferase mRNA and dependency on GAL4 gene function. Proc. Natl Acad. Sci. USA 75, 2878–2882 (1978).
Dultz, E. et al. Global reorganization of budding yeast chromosome conformation in different physiological conditions. J. Cell Biol. 212, 321–334 (2016).
Shechtman, Y. et al. Observation of live chromatin dynamics in cells via 3D localization microscopy using Tetrapod point spread functions. Biomed. Opt. Express 8, 5735 (2017).
Sahay, G. et al. Efficiency of siRNA delivery by lipid nanoparticles is limited by endocytic recycling. Nat. Biotechnol. 31, 653–658 (2013).
Veiga, N. et al. Cell specific delivery of modified mRNA expressing therapeutic proteins to leukocytes. Nat. Commun. 9, 4493 (2018).
van der Meel, R. et al. Smart cancer nanomedicine. Nat. Nanotechnol. 14, 1007–1017 (2019).
Tsang, M., Gantchev, J., Ghazawi, F. M. & Litvinov, I. V. Protocol for adhesion and immunostaining of lymphocytes and other non-adherent cells in culture. Biotechniques 63, 230–233 (2017).
Shechtman, Y., Sahl, S. J., Backer, A. S. & Moerner, W. E. Optimal point spread function design for 3D imaging. Phys. Rev. Lett. 113, 133902 (2014).
Nehme, E. et al. DeepSTORM3D: dense three dimensional localization microscopy and point spread function design by deep learning. Preprint at https://arxiv.org/abs/1906.09957 (2019).
Boyd, N., Jonas, E., Babcock, H. P. & Recht, B. DeepLoco: fast 3D localization microscopy using neural networks. Preprint at https://doi.org/10.1101/267096 (2018).
Nitta, N. et al. Intelligent image-activated cell sorting. Cell 175, 266–276.e13 (2018).
Egner, A. & Hell, S. W. in Handbook of Biological Confocal Microscopy 3rd edn (eds Egener, A. & Hell, S.W.) 404–413 (Springer, 2006).
Jia, S., Vaughan, J. C. & Zhuang, X. Isotropic three-dimensional super-resolution imaging with a self-bending point spread function. Nat. Photonics 8, 302–306 (2014).
Backer, A. S., Backlund, M. P., Von Diezmann, A. R., Sahl, S. J. & Moerner, W. E. A bisected pupil for studying single-molecule orientational dynamics and its application to three-dimensional super-resolution microscopy. Appl. Phys. Lett. 104, 193701 (2014).
Bergman, L. W., Saghbini, M., Hoekstra, D. & Gautsch, J. Growth and maintenance of yeast. Media formulations for various two-hybrid systems. Methods Mol. Biol. 177, 15–39 (2001).
Hansen, A. S., Hao, N. & O’Shea, E. K. High-throughput microfluidics to control and measure signaling dynamics in single yeast cells. Nat. Protoc. 10, 1181–1197 (2015).
Sela, M. et al. Sequential phosphorylation of SLP-76 at tyrosine 173 is required for activation of T and mast cells. EMBO J. 30, 3160–3172 (2011).
Linkert, M. et al. Metadata matters: access to image data in the real world. J. Cell Biol. 189, 777–782 (2010).
Schindelin, J. et al. Fiji: an open-source platform for biological-image analysis. Nat. Methods 9, 676–682 (2012).
Ovesný, M., Křížek, P., Borkovec, J., Svindrych, Z. & Hagen, G. M. ThunderSTORM: a comprehensive ImageJ plug-in for PALM and STORM data analysis and super-resolution imaging. Bioinformatics 30, 2389–2390 (2014).
Acknowledgements
We thank E. Barak, Y. Lupu-Haber, M. Duvshani-Eshet and the Lorry I. Lokey Interdisciplinary Center for Life Sciences and Engineering for technical assistance with the imaging flow cytometer. The multicolour yeast cells were provided by K. Weis and E. Dultz and the Jurkat human T lymphocytes were provided by D. Yablonski, and verified with the assistance of the Genomic Center at the Technion Biomedical Core Facility. The Tetrapod phase mask was fabricated by M.Y. Lee. This work was partially supported by the European Research Council (ERC) under the European Union Horizon 2020 research and innovation programme to Y.S. (grant no. 802567), and an ERC Starting Grant to A.S. (ERC-STG-2015–680242), the Zuckerman foundation, the POLAK Fund for Applied Research at the Technion and the Israel Science Foundation (1421/17) to A.S. and (450/18) to Y.S.
Author information
Authors and Affiliations
Contributions
L.E.W. and Y.S. conceived of the approach. L.E.W., Y.S.E., O. Adir, A.S. and Y.S. designed the experiments. L.E.W., Y.S.E., S.G., O. Adir, B.F. and O. Alalouf performed the experiments. All authors contributed to data analysis and preparing the manuscript.
Corresponding authors
Ethics declarations
Competing interests
L.E.W. and Y.S. are inventors on a patent application (International Publication no. WO2019180705A1) concerning the described technology.
Additional information
Peer review information Nature Nanotechnology thanks Jörg Enderlein, Tom Misteli and the other anonymous reviewer(s) for their contribution to the peer review of this work.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary Figs. 1–8, figure captions, discussion and sample preparation details.
Rights and permissions
About this article
Cite this article
Weiss, L.E., Shalev Ezra, Y., Goldberg, S. et al. Three-dimensional localization microscopy in live flowing cells. Nat. Nanotechnol. 15, 500–506 (2020). https://doi.org/10.1038/s41565-020-0662-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41565-020-0662-0
This article is cited by
-
Light-field flow cytometry for high-resolution, volumetric and multiparametric 3D single-cell analysis
Nature Communications (2024)
-
3D printable diffractive optical elements by liquid immersion
Nature Communications (2021)
-
Axial localization and tracking of self-interference nanoparticles by lateral point spread functions
Nature Communications (2021)
-
Single-molecule imaging goes high throughput
Nature Nanotechnology (2020)
-
Recent advances in point spread function engineering and related computational microscopy approaches: from one viewpoint
Biophysical Reviews (2020)