Abstract
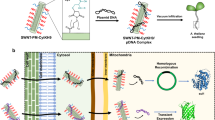
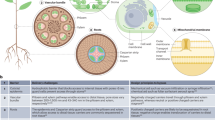
Plant genetic engineering is an important tool used in current efforts in crop improvement, pharmaceutical product biosynthesis and sustainable agriculture. However, conventional genetic engineering techniques target the nuclear genome, prompting concerns about the proliferation of foreign genes to weedy relatives. Chloroplast transformation does not have this limitation, since the plastid genome is maternally inherited in most plants, motivating the need for organelle-specific and selective nanocarriers. Here, we rationally designed chitosan-complexed single-walled carbon nanotubes, utilizing the lipid exchange envelope penetration mechanism. The single-walled carbon nanotubes selectively deliver plasmid DNA to chloroplasts of different plant species without external biolistic or chemical aid. We demonstrate chloroplast-targeted transgene delivery and transient expression in mature Eruca sativa, Nasturtium officinale, Nicotiana tabacum and Spinacia oleracea plants and in isolated Arabidopsis thaliana mesophyll protoplasts. This nanoparticle-mediated chloroplast transgene delivery tool provides practical advantages over current delivery techniques as a potential transformation method for mature plants to benefit plant bioengineering and biological studies.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Data availability
The data that support the plots within this paper and other findings of this study are available from the corresponding author upon reasonable request.
References
Wang, Y. et al. Simultaneous editing of three homoeoalleles in hexaploid bread wheat confers heritable resistance to powdery mildew. Nat. Biotechnol. 32, 947–951 (2014).
Abdallah, N. A., Prakash, C. S. & McHughen, A. G. Genome editing for crop improvement: challenges and opportunities. GM Crops Food 6, 183–205 (2015).
Marsian, J. et al. Plant-made polio type 3 stabilized VLPs—a candidate synthetic polio vaccine. Nat. Commun. 8, 245 (2017).
Li, J. F. et al. Multiplex and homologous recombination-mediated genome editing in Arabidopsis and Nicotiana benthamiana using guide RNA and Cas9. Nat. Biotechnol. 31, 688–691 (2013).
Yin, K., Gao, C. & Qiu, J.-L. Progress and prospects in plant genome editing. Nat. Plants 3, 17107 (2017).
Duke, S. O. Perspectives on transgenic, herbicide-resistant crops in the United States almost 20 years after introduction. Pest. Manag. Sci. 71, 652–657 (2015).
Fischer, R., Stoger, E., Schillberg, S., Christou, P. & Twyman, R. M. Plant-based production of biopharmaceuticals. Curr. Opin. Plant Biol. 7, 152–158 (2004).
Fuentes, P., Armarego-Marriott, T. & Bock, R. Plastid transformation and its application in metabolic engineering. Curr. Opin. Biotechnol. 49, 10–15 (2018).
Jin, S. & Daniell, H. The engineered chloroplast genome just got smarter. Trends Plant Sci. 20, 622–640 (2015).
Maliga, P. in Genomics of Chloroplasts and Mitochondria (Advances in Photosynthesis and Respiration Series, Vol. 35) 393–414 (Springer, Dordrecht, 2012).
Scott, S. E. & Wilkinson, M. J. Low probability of chloroplast movement from oilseed rape (Brassica napus) into wild Brassica rapa. Nat. Biotechnol. 17, 390–392 (1999).
De Cosa, B., Moar, W., Lee, S. B., Miller, M. & Daniell, H. Overexpression of the Bt cry2Aa2 operon in chloroplasts leads to formation of insecticidal crystals. Nat. Biotechnol. 19, 71–74 (2001).
Staub, J. M. et al. High-yield production of a human therapeutic protein in tobacco chloroplasts. Nat. Biotechnol. 18, 333–338 (2000).
Svab, Z. & Maliga, P. High-frequency plastid transformation in tobacco by selection for a chimeric aadA gene. Proc. Natl Acad. Sci. USA 90, 913–917 (1993).
Golds, T., Maliga, P. & Koop, H. U. Stable plastid transformation in PEG-treated protoplasts of Nicotiana tabacum. Nat. Biotechnol. 11, 95–97 (1993).
Cunningham, F. J., Goh, N. S., Demirer, G. S., Matos, J. L. & Landry, M. P. Nanoparticle-mediated delivery towards advancing plant genetic engineering. Trends Biotechnol. 36, 882–897 (2018).
Rafsanjani, M. S. O., Alvari, A., Samim, M., Hejazi, M. A. & Abdin, M. Z. Application of novel nanotechnology strategies in plant biotransformation: a contemporary overview. Recent Pat. Biotechnol. 6, 69–79 (2012).
Ahmad, N., Michoux, F., Lössl, A. G. & Nixon, P. J. Challenges and perspectives in commercializing plastid transformation technology. J. Exp. Bot. 67, 5945–5960 (2016).
Whitehead, K. A. et al. Degradable lipid nanoparticles with predictable in vivo siRNA delivery activity. Nat. Commun. 5, 4277 (2014).
Wang, H. et al. Biocompatible chitosan–carbon dot hybrid nanogels for NIR-imaging-guided synergistic photothermal–chemo therapy. ACS Appl. Mater. Interfaces 9, 18639–18649 (2017).
Deng, X. et al. Hyaluronic acid-chitosan nanoparticles for co-delivery of MiR-34a and doxorubicin in therapy against triple negative breast cancer. Biomaterials. 35, 4333–4344 (2014).
Tripathi, D. K. et al. An overview on manufactured nanoparticles in plants: uptake, translocation, accumulation and phytotoxicity. Plant Physiol. Biochem. 110, 2–12 (2017).
Torney, F., Trewyn, B. G., Lin, V. S. Y. & Wang, K. Mesoporous silica nanoparticles deliver DNA and chemicals into plants. Nat. Nanotechnol. 2, 295–300 (2007).
Giraldo, J. P. et al. Plant nanobionics approach to augment photosynthesis and biochemical sensing. Nat. Mater. 13, 400–408 (2014).
Wong, M. H. et al. Lipid exchange envelope penetration (LEEP) of nanoparticles for plant engineering: a universal localization mechanism. Nano Lett. 16, 1161–1172 (2016).
Lew, T. T. S. et al. Rational design principles for the transport and subcellular distribution of nanomaterials into plant protoplasts. Small 14, 1802086 (2018).
Li, Z., de Barros, A. L. B., Soares, D. C. F., Moss, S. N. & Alisaraie, L. Functionalized single-walled carbon nanotubes: cellular uptake, biodistribution and applications in drug delivery. Int. J. Pharm. 524, 41–54 (2017).
Liu, Q. et al. Carbon nanotubes as molecular transporters for walled plant cells. Nano Lett. 9, 1007–1010 (2009).
Wu, Y., Phillips, J. A., Liu, H., Yang, R. & Tan, W. Carbon nanotubes protect DNA strands during cellular delivery. ACS Nano 2, 2023–2028 (2008).
Malerba, M. & Cerana, R. Recent advances of chitosan applications in plants. Polymers 10, 118 (2018).
Choudhary, R. C. et al. Cu-chitosan nanoparticle boost defense responses and plant growth in maize (Zea mays L.). Sci. Rep. 7, 9754 (2017).
Shearer, C. J. et al. Adsorption and desorption of single-stranded DNA from single-walled carbon nanotubes. Chem. Asian J. 12, 1625–1634 (2017).
Yang, Y. et al. Binding efficacy and kinetics of chitosan with DNA duplex: the effects of deacetylation degree and nucleotide sequences. Carbohydr. Polym. 169, 451–457 (2017).
Jokerst, J. V., Lobovkina, T., Zare, R. N. & Gambhir, S. S. Nanoparticle PEGylation for imaging and therapy. Nanomedicine 6, 715–728 (2011).
Mathur, J. & Koncz, C. in Arabidopsis Protocols (Methods in Molecular Biology Series, Vol. 82) 267–276 (Humana, Totowa, 1998).
Fettiplace, R., Andrews, D. M. & Haydon, D. A. Thickness, composition and structure of some lipid bilayers and natural membranes. J. Membr. Biol. 5, 277–296 (1971).
Zimmermann, U. & Neil, G. A. Electromanipulation of Cells (CRC, Boca Raton, 1996).
Heikkila, E. et al. Cationic Au nanoparticle binding with plasma membrane-like lipid bilayers: potential mechanism for spontaneous permeation to cells revealed by atomistic simulations. J. Phys. Chem. C 118, 11131–11141 (2014).
Wang, B., Zhang, L., Bae, S. C. & Granick, S. Nanoparticle-induced surface reconstruction of phospholipid membranes. Proc. Natl Acad. Sci. USA 105, 18171–18175 (2008).
Kupiainen, M. et al. Free volume properties of sphingomyelin, DMPC, DPPC, and PLPC bilayers. J. Comput. Theor. Nanosci. 2, 401–413 (2005).
Alberts, B. et al. Molecular Biology of the Cell 4th edn (Garland Science, New York, 2002).
Mao, S., Sun, W. & Kissel, T. Chitosan-based formulations for delivery of DNA and siRNA. Adv. Drug Deliv. Rev. 62, 12–27 (2010).
Kruss, S. et al. Neurotransmitter detection using corona phase molecular recognition on fluorescent single-walled carbon nanotube sensors. J. Am. Chem. Soc. 136, 713–724 (2014).
Yang, R. et al. Carbon nanotube-quenched fluorescent oligonucleotides: probes that fluoresce upon hybridization. J. Am. Chem. Soc. 130, 8351–8358 (2008).
Lesney, M. S. Polycation-like behaviour of chitosan on suspension-culture derived protoplasts of slash pine. Phytochemistry 29, 1123–1125 (1990).
Kwak, S. Y. et al. A nanobionic light-emitting plant. Nano Lett. 17, 7951–7961 (2017).
Díaz, A. H. & Koop, H.-U. in Chloroplast Biotechnology (Methods in Molecular Biology Series, Vol. 1132) 165–175 (Humana, Totowa, 2014).
Ruhlman, T. A. in Chloroplast Biotechnology (Methods in Molecular Biology Series, Vol. 1132) 331–343 (Humana, Totowa, 2014).
Asensio, J. L., Ardá, A., Cañada, F. J. & Jiménez-Barbero, J. Carbohydrate–aromatic interactions. Acc. Chem. Res. 46, 946–954 (2013).
Suzuki, J. Y., Sriraman, P., Svab, Z. & Maliga, P. Unique architecture of the plastid ribosomal RNA operon promoter recognized by the multisubunit RNA polymerase in tobacco and other higher plants. Plant Cell 15, 195–205 (2003).
Bohmert-Tatarev, K., McAvoy, S., Daughtry, S., Peoples, O. P. & Snell, K. D. High levels of bioplastic are produced in fertile transplastomic tobacco plants engineered with a synthetic operon for the production of polyhydroxybutyrate. Plant Physiol. 155, 1690–1708 (2011).
Thompson, C. J. et al. Characterization of the herbicide-resistance gene bar from Streptomyces hygroscopicus. EMBO J. 6, 2519–2523 (1987).
Kuroda, H. & Maliga, P. Sequences downstream of the translation initiation codon are important determinants of translation efficiency in chloroplasts. Plant Physiol. 125, 430–436 (2001).
Herz, S., Füßl, M., Steiger, S. & Koop, H. U. Development of novel types of plastid transformation vectors and evaluation of factors controlling expression. Transgenic Res. 14, 969–982 (2005).
Jefferson, R. A., Kavanagh, T. A. & Bevan, M. W. GUS fusions: beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J. 6, 3901–3907 (1987).
Staub, J. M. & Maliga, P. Accumulation of D1 polypeptide in tobacco plastids is regulated via the untranslated region of the psbA mRNA. EMBO J. 12, 601–606 (1993).
Benfey, P. N. & Chua, N. H. Regulated genes in transgenic plants. Science 244, 174–181 (1989).
van der Krol, A. R. & Chua, N. H. The basic domain of plant B-ZIP proteins facilitates import of a reporter protein into plant nuclei. Plant Cell 3, 667–675 (1991).
Yoo, S. D., Cho, Y. H. & Sheen, J. Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nat. Protoc. 2, 1565–1572 (2007).
Littlejohn, G. R., Gouveia, J. D., Edner, C., Smirnoff, N. & Love, J. Perfluorodecalin enhances in vivo confocal microscopy resolution of Arabidopsis thaliana mesophyll. New Phytol. 186, 1018–1025 (2010).
Ueki, S., Lacroix, B., Krichevsky, A., Lazarowitz, S. G. & Citovsky, V. Functional transient genetic transformation of Arabidopsis leaves by biolistic bombardment. Nat. Protoc. 4, 71–77 (2009).
Acknowledgements
This research was supported by the National Research Foundation (NRF), Prime Minister’s Office, Singapore under its Campus for Research Excellence and Technological Enterprise (CREATE) program. The Disruptive & Sustainable Technology for Agricultural Precision (DiSTAP) is an interdisciplinary research group (IRG) of the Singapore MIT Alliance for Research and Technology (SMART) Centre. We also acknowledge support of Sime Darby Malaysia. T.T.S.L. and M.H.W. were supported on a graduate fellowship by the Agency of Science, Research and Technology, Singapore. V.B.K. was supported by The Swiss National Science Foundation (project no. P300P2_174469). The authors are grateful for helpful discussion with J. P. Giraldo and thank M. M. R. Ambavaram of Yield10 Bioscience for technical guidance and C. Xu of Yield10 Bioscience (now at Jounce Therapeutics) for technical assistance.
Author information
Authors and Affiliations
Contributions
S.-Y.K. and T.T.S.L. co-wrote the paper. S.-Y.K., T.T.S.L. and M.S.S. conceived and designed the experiments. C.J.S. assisted with the preparation and characterization of the chitosan-complexed SWNTs. V.B.K. performed AFM analysis. M.H.W. assisted with experimental design. K.D.S., K.B.-T. and J.S.S. constructed plasmid DNA. K.D.S. and K.B.-T. contributed the plastid DNA construct pMBX1120. J.S.S. and N.-H.C. constructed the nuclear DNA construct pBA-GFP-NL. All authors have revised the manuscript and given their approval of the final version.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary Figures 1–11
Rights and permissions
About this article
Cite this article
Kwak, SY., Lew, T.T.S., Sweeney, C.J. et al. Chloroplast-selective gene delivery and expression in planta using chitosan-complexed single-walled carbon nanotube carriers. Nat. Nanotechnol. 14, 447–455 (2019). https://doi.org/10.1038/s41565-019-0375-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41565-019-0375-4
This article is cited by
-
Construction of peptide/plasmid DNA complexes for plant gene transfection via the basic leucine zipper domain
Polymer Journal (2024)
-
An Overview of Targeted Genome Editing Strategies for Reducing the Biosynthesis of Phytic Acid: an Anti-nutrient in Crop Plants
Molecular Biotechnology (2024)
-
Proteomic insights to decipher nanoparticle uptake, translocation, and intercellular mechanisms in plants
Environmental Science and Pollution Research (2024)
-
Application of nanotechnology and proteomic tools in crop development towards sustainable agriculture
Journal of Crop Science and Biotechnology (2024)
-
Nanomaterials for intelligent CRISPR-Cas tools: improving environment sustainability
Environmental Science and Pollution Research (2024)