Abstract
Although repetitive patterns of antigens are crucial for certain immune responses, an understanding of how antibodies bind and dynamically interact with various spatial arrangements of molecules is lacking. Hence, we introduced a new method in which molecularly precise nanoscale patterns of antigens are displayed using DNA origami and immobilized in a surface plasmon resonance set-up. Using antibodies with identical antigen-binding domains, we found that all the subclasses and isotypes studied bind bivalently to two antigens separated at distances that range from 3 to 17 nm. The binding affinities of these antibodies change with the antigen distances, with a distinct preference for antigens separated by approximately 16 nm, and considerable differences in spatial tolerance exist between IgM and IgG and between low- and high-affinity antibodies.
Similar content being viewed by others
Main
Rigidly organized molecular patterns are typically either foreign or intracellular in most mammals and the immune system has thus evolved an efficient response to such structures1. In fact, the concept of particulate antigen display has been successfully exploited for vaccine development2,3 (Supplementary Note 1). Exactly how such patterns elicit a strong response is, however, still not fully understood. A recent study using high-speed atomic force microscopy suggested that antibodies, and thus also, by extension, B-cell receptors (BCRs), are able to ‘walk’ on a pattern of antigens by attachment and release of its Fab arms4. This concept, combined with a clustering model of BCR activation5, could provide more clues as to why B-cell activation is strong for rigid and highly organized patterns of antigens. Whatever the case may be, a picture is nevertheless emerging in which it is becoming clear that a deeper knowledge of the dynamics of antibody binding to variably distributed antigens would be highly desirable for a more complete understanding of the initiation of immune responses and for rational vaccine design.
Multivalent binding interaction between antibodies and antigens is also regarded as one of the dominant modes to initiate antibody effector functions (Supplementary Note 1), such as complement activation6, antibody-dependent cell-mediated cytotoxicity7, opsonophagocytosis and antibody-mediated antigen presentation8,9. Thus, the geometric organization of antigens not only plays an important role in influencing initiation but also on the induced antibody isotype and subclass effector functions.
Antibodies consist of two identical antigen binding fragment arms (Fab) and a constant fragment crystallizable region (Fc) (Fig. 1a and Supplementary Fig. 1) in a homodimeric molecule connected via disulfide bridges10. Humans have five antibody isotypes (IgM, IgD, IgG, IgE and IgA) and IgG, the most abundant in plasma, can further be divided into four subclasses (IgG1, IgG2, IgG3 and IgG4). Sequence differences between the antibody isotypes (Supplementary Figs. 1 and 2) result in several differences, for example, the number of domains of their heavy chains and the makeup of the flexible hinge region located between the two Fabs and the Fc region. The human IgG subclasses have minor differences in amino acid composition in the Fc region, but have substantial differences in amino acid sequence and length of their hinge regions. These differences result in very distinct functional properties and engagement of Fc receptors and the complement system11. Crystal structures of full-length IgG are few10 (Supplementary Fig. 1) due to their flexibility6,12, but have revealed strikingly asymmetric conformations somewhere between a T and a Y shape. Electron microscopy has been used to estimate that IgG should be capable of bivalently binding antigens separated by 6–14 nm for the mouse13 or human14, whereas fluorescence energy transfer studies suggest a slightly wider bivalent binding distance15.
a, Three-dimensional (3D) rendering of human IgG1 based on X-ray crystallography data (Protein Data Bank PDB:1HZH)10. b, The advantage of PSPR: in contrast to conventional SPR (left), which randomly arranges its ligands on the surface, the PSPR method (right) utilizes DNA origami to prepattern the antigen of interest (yellow dots) prior to immobilization. c, Antigen nanopatterns were fabricated using different combinations of antigen-decorated staple oligonucleotides (differently coloured lines). d, 3D models using cylinders as a representation of double helices (left) and transmission electron microscopy negative-stain micrographs (right) of the DNA nanostructures used in this study. Scale bars, 40 nm. Two types of DNA nanostructures, an 18-helix rod and a 44-helix brick, were used to pattern the antigens. e, The antigen nanopatterns were immobilized onto a streptavidin-biotinylated oligonucleotide surface via oligonucleotide hybridization to sequences that protrude from the bottom of the origami. f,g, These were first allowed to immobilize in the SPR machine (f), followed by an injection of increasing antibody concentrations and finally a dissociation phase (g). The kinetic data can be obtained by fitting the binding curves with a 1:1 binding model (Supplementary Fig. 7). The model shows the origami and antibody rendered to scale, and illustrates a 16 nm bivalent binding and a monovalent binding.
Despite all the clues to its importance, the answer to the question of how structural flexibility affects the ability of antibodies to bind their cognate antigens at different distances and densities remains largely unknown. In particular, no methods have been able to show precisely the optimal antigen separation required for the most stable bivalent bindings.
Design and characterization of PSPR
To characterize the dynamic interplay between the antibody structural flexibility and binding to cognate antigen patterns in real time, we applied DNA nanotechnology16 using DNA origami17,18,19, which is used to arrange proteins or chemicals such as fluorophores, with nanometre20,21,22,23 or subnanometre precision24, including demonstrations of bivalent binding25. The method was established by the conjugation of hapten (a small-molecule antigen) patterns to well-defined DNA origami nanostructures (Fig. 1b–d) that were immobilized on a surface plasmon resonance (SPR) chip using oligonucleotide hybridization (Fig. 1e,f). We designed two DNA origami nanostructures, an 18-helix bundle (18HB) and a brick used for pairwise distance measurements or multiantigen 2D patterns, respectively (Fig. 1d, Supplementary Figs. 3–5 and Supplementary Table 1) patterned with either digoxigenin (DIG), 4-hydroxy-3-iodo-5-nitrophenylacetate (NIP) or 4-hydroxy-3-nitrophenyl (NP) antigens. The repertoire of human anti-NIP antibodies with equivalent Fab affinities established previously (Methods) constitutes an opportunity to study and compare medically relevant classes of antibodies and was used for the main experiments as far as possible with NP or NIP as the antigens. To establish the method, and in particular when strong monovalent binding affinity was needed, as in the case of complex binding patters, we resorted to the use of commercially available high-affinity rabbit anti-DIG IgG. Serial dilutions of anti-hapten antibodies were injected over stably immobilized DNA nanostructures and the obtained sensorgrams (single-cycle kinetics) (Fig. 1g) were fitted to a 1:1 Langmuir binding model to derive the binding constants (Supplementary Figs. 6 and 7). We refer to the method as patterned SPR (PSPR).
To validate the robustness of the PSPR method, we took advantage of monoclonal rabbit anti-DIG IgG antibodies (alignment data in Supplementary Figs. 8 and 9) and measured binding to origami patterns with DIG. The structures that displayed widely spaced antigens induced by monovalent binding resulted in binding affinities in the apparent dissociation constant (Kd) of 25–35 pM, where the binding occupancy scaled well with the number of antigens per structure (Supplementary Fig. 10; the binding kinetics data are given in Supplementary Table 2). ANOVA analysis showed that the location of the antigens on the monovalent nanostructure did not affect the binding interaction significantly (Supplementary Table 3). Nanostructures with two closely spaced hapten molecules showed a significant increase in binding affinity (~tenfold decrease in Kd), with a binding occupancy of one antibody per structure, consistent with the formation of bivalent binding. Nanostructures with three antigens gained binding affinities between monovalent and bivalent binding, in which the binding occupancy resulted in roughly two antibodies per structure, indicating a mixture of monovalent and bivalent binding. Furthermore, we introduced some extra mobility to the antigens by increasing the linker length from 2 to 21 nucleotides, which resulted in similar antibody binding patterns (Supplementary Fig. 11). Removing the 2-nucleotide linker gave no significant difference in the antibody’s binding affinity towards the 3.4 nm sample, which suggests that the uncertainty of distances between two antigens is smaller than the linker’s contour length, probably due to the entropic spring-like behaviour of both the linkers and the overall structural flexibility (Supplementary Fig. 12 and Methods). Small molecules displayed on origami constitute state-of-the-art molecular positioning, so, although distances can vary from fluctuations in individual structures, the ensemble average nature of our measurements over a large number of entropic springs ensures robust comparisons over nanometre distances, similar to previous studies using origami distance measurements24,25. As the density of individual nanostructures on the SPR chip may influence the binding kinetics by allowing a single antibody to bridge two such neighbouring structures, we verified that the seeding density in our case did not influence the binding affinity for the concentrations of the structures used (Supplementary Fig. 13). Quantitative incorporation of hapten-modified staples to the nanostructures is also key to the PSPR method. Using gel retardation assays, we demonstrated that an undetectable amount of our nanostructures have defects in hapten oligonucleotide incorporation (Supplementary Figs. 4 and 14).
Exploring the structural flexibility of human IgG subclasses
Next, we aimed to explore the conformational flexibility of the four human IgG subclasses by the use of mouse–human chimeric versions that harbour specificity for the hapten NIP (Methods). Binding was studied using the 18HB on which two NIP molecules were arranged at distances from 3 to 44 nm (Supplementary Fig. 5) and a monovalently decorated structure (denoted ‘0 nm’) (Fig. 2 and Supplementary Table 4). We observed bivalent binding as a significant decrease (>1,500-fold) in the Kd for optimal distances compared to the monovalent samples. Interestingly, instead of a sharp cut-off between monovalent and bivalent binding, a U-shaped behaviour in the dissociation constant, which corresponded to a sharp peak in association, was seen when the binding affinity was plotted against antigen distance (Fig. 2 and Supplementary Figs. 15, 16). Binding became weaker when two antigens were arranged at short (3 nm) or long (17 nm) separations, and the binding affinity reached a striking maximum at 16 nm for all the antibodies studied. Monovalent binding clearly resumed at 29 nm. Importantly, the results revealed differences between the IgG subclasses, where IgG1 and IgG4 showed a similar behaviour, but IgG2, which is the most rigid subclass, showed a ‘stepwise’ behaviour, in which the binding affinity was similar at short distances but became significantly stronger at around 14–16 nm. In contrast, IgG3, which is the most flexible subclass, showed strong binding at shorter distances and even more so at 14–16 nm. Surprisingly, although the strongest binding was seen at 16 nm, there was a sharp drop in affinity when going to 17 nm (~tenfold increase in Kd), which indicates that this distance range of around 16 nm is both optimal and at the same time close to the limiting distance for bivalent binding in the four human IgG subclasses. An illustration of IgG1 stretched to a 16 nm bivalent binding is shown in Fig. 1g.
Human anti-NIP antibodies binding to two NIP molecules patterned on the 18HB at varying separations. a,b, Scatter graphs of Kd and binding ratio (maximum resonance unit (RU) of antibodies/RU of nanostructures, which corresponds to average antibodies per structure) for the various separations. Exact structure designs are given in d and Supplementary Fig. 5. The ‘0’ distance is just one displayed antigen. The y axis (top panels) is the apparent binding affinity recorded and fitted with BIAcore t200 evaluation software. The amount of antibody (in RU) that is bound to 1 RU of the structure (the binding ratio) is plotted in the bottom panels. a, Data for all wild type IgG subclasses. b, Data for IgG3 hinge mutants and monomeric IgM. y-axis error bars = s.e.m., sometimes smaller than the markers; for samples without error bars, n = 1. Distance (x axis) error bars are the s.d. from a uniform ±2 nm distribution added to a 3% s.d. derived from structure fluctuations of the 18HB (Methods). c, The same data as in a plotted as association constants, Ka (= 1/Kd), versus separations for the IgG subclasses. A peak behaviour in the changes of Ka was observed for all the tested antibodies, the bivalent binding becomes weaker at short (3–7 nm) and long (17 nm) distances, and this behaviour varies between different antibody subclasses (n = 3 or 4 (detailed description in Supplementary Table 4); central values = average value). d, Illustrations (top (left) and perspective (right) views) of the 18HB show the locations of the NIP-modified staple-oligonucleotides in blue and the NIP locations highlighted with white arrowheads. The distances are shown as the design distance ± distance error calculated as described in Methods.
To further explore the antigen binding properties of IgG3, and in particular the influence of its long hinge of 62 amino acids, we studied three hinge-engineered variants; one variant harboured only the C-terminal part of the hinge, which consisted of 15 amino acids (m15), one in which the hinge was replaced with a short stretch of five amino acids (Ala-Ala-Ala-Cys-Ala), which included a cysteine that made a disulfide bridge between the heavy chains (HM4), and one that lacked the hinge region altogether, such that the heavy chains were held together solely by non-covalent interactions in the CH3 domains and the CH2 domains are linked by a disulfide bridge at the C-terminal end of the light chains (HM5) (Supplementary Fig. 17). The results revealed that the HM4 mutant exhibited an IgG2-like behaviour, with a strong preference for bivalent binding to antigens spaced in the 14–16 nm range, whereas the m15 mutant showed a binding behaviour similar to that of IgG1 and IgG4. This result is in line with what one would expect, given the length of the upper hinge and thus the flexibility of these mutants. Interestingly, IgG3 and both the HM4 and HM5 mutants showed a peculiar behaviour with a local minimum in Kd around 3 nm. Importantly, we studied the binding behaviour of an engineered monomeric IgM variant (C575S), which, unlike its natural counterpart, does not form hexamers and/or pentamers. IgM does not contain a hinge region, but instead has an additional domain (Supplementary Fig. 1). Despite lacking a hinge region, we observed strong bivalent binding for antigen separations in the range 3–17 nm (Fig. 2b and Supplementary Fig. 18), and the relatively low Kd at 29 nm (which shows similar affinities as the IgGs displayed for 17 nm) indicates that, to some extent, monomeric IgM is either able to stretch, perhaps by partial unfolding of the domains for bivalent binding well beyond 17 nm, or is able to multimerize to reach long distances effectively (which would be consistent with the higher binding ratio seen in Fig. 2b). This remarkable ability of monomeric IgM to bind bivalently to antigens spaced over a wide range of distances, or spatial tolerance, may well be crucial for its special role in the first line of defence against pathogens as the IgM BCR is monomeric on the surface of B cells.
Next, we explored the relationship between the binding affinity of the individual Fab and conformational flexibility of the anti-NIP antibodies using the hapten NP as the antigen instead of NIP, which gave a 60-fold weaker Kd than that measured towards NIP conjugated nanostructures. We observed that the antibodies failed to crosslink two NP antigens by bivalent binding at all distances tested, except the 16 nm distance, for both IgG1 and IgG3 (Fig. 3). Thus, lower affinity antibodies clearly have a lower spatial tolerance than the stronger binders. Note that the antibodies were identical (two different antigens were used) and flexibility arguments cannot explain the data, but instead suggest that the lower binding energy of the individual Fab arms are not strong enough to induce stretching or contraction of the arms, as may well be the case when binding to NIP.
A comparison of human anti-NIP IgG1 and IgG3 binding to NIP (right y axis) or NP (left y axis). The separation distance is between two antigens. See Fig. 2 and Supplementary Fig. 5 for exact structure designs. The point at 0 represents a structure with one antigen. NIP data points are identical to those of Fig. 2, and their binding affinities (blue) correspond to the right y axis. The NP binding affinity (Kd) (red) is plotted on the left y axis. (n = 3; central values = average value; error bars = s.e.m. For samples without error bars, n = 1.)
Deconstruction of multiple antibody–antigen interactions
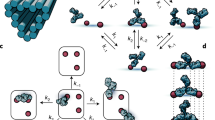
To deconstruct the complex interactions between multiple antibodies and antigens, we designed PSPR experiments with nanoscale patterns of DIG monomers, dimers, trimers, tetramers, pentamers and hexamers (brick structures, H1–H6 hexagon with 15 nm edges (Fig. 4a and Supplementary Fig. 5)) with the aim to construct and empirically calibrate a model of multivalent binding dynamics. We measured both the apparent affinity and average binding ratio (corresponding to average antibodies per structure) using rabbit IgG (Fig. 4a and data in Supplementary Table 5). Notably, although the tetramers and hexamers were engineered to enable a saturated state of bivalently bound antibodies, we observed greater binding ratios than would be expected if this was the case. In contrast to single molecule methods, which have been employed to construct models of molecular dynamics according to individual trajectories26, we implemented a mathematical model that dissects the ensemble PSPR signal (four-parameter continuous-time Markov chain) and was able to accurately reproduce the binding curves from a bottom-up quantification of the contributions of internal molecular states (Fig. 4a–c). This dynamic model (detailed in Supplementary Note 2 and Supplementary Figs. 19–31) assumes that the local states are unoccupied, monovalently occupied and bivalently occupied, and a network of transitions between those states mediated is by the elementary processes of monovalent attachment, monovalent detachment, monovalent-to-bivalent conversion and bivalent-to-monovalent conversion. We sought a consensus set of the four rate parameters with which our model was able to produce satisfactory fits for all the partial configurations of the hexamer antigen patterns (Supplementary Fig. 23). Compared to previous bivalent binding models27 that strive to accurately reproduce the dynamics of multivalent binding, our model can be used to predict the dynamics of binding to hypothetical antigen patterns of arbitrary geometry and provides data on internal conversion rates (Fig. 4c–e) and constituent microstates (Fig. 4b–e).
a, Comparison of final measured binding ratio (orange) and apparent Kd values (blue) determined for different nanopatterns (top row: H, hexagon; number of antigens displayed; c, close; f, far) of DIG on the brick structure (top left) in which the antigens are displayed in a regular hexagon with edge distances of approximately 15 nm (Fig. 1 and Supplementary Fig. 5) using rabbit IgG (n = 3; central values = average value; error bars = s.d.). b, Their corresponding most-dominant microstates determined by a model of steady-state binding behaviour. c, Example of an SPR curve (cyan) for a partial hexamer configuration (H2c in a) and corresponding fitted model output (magenta). Inset: schematic visualization of the network of states for this system and transitions between those states governed by the four rate parameters. d, Weighted probability trajectory for the hexameric antigen pattern. Each state has a corresponding occupancy, which can be used to weight its probability according to its contribution to the SPR signal. The stacked summation of all the weighted probabilities reproduces the SPR curve for that experiment. The equation describes the total occupancy (O) as the expectation value function of individual state occupancies. Inset: the unweighted probability distribution of states, including the empty state initial condition in blue. e, Rate constants, which include internal monovalent–bivalent kinetics, determined by model fitting for rabbit IgG using data from binding to the brick patterns. Ab, antibodies.
Hexameric antigens are of particular interest as self-assembled Fc–Fc-formed antibody hexamers that are monovalently bound to their antigens have been demonstrated to activate the complement system with a high efficiency6. Our results show that the dominant interaction between an antibody and antigen hexamer patterns with 15 nm separations is the engagement of the bivalent binding of three antibodies, and the probability of having six monovalently bound antibodies is close to zero even at a 0.5 nM antibody concentration (530 ppm among all the possible microstates). However, the model of binding dynamics indicates that at the steady state, antibodies in every system assume a diverse distribution of states, dominated by bivalent saturation configurations (for example, three bivalently bound antibodies in the hexameric configuration). The probability space is shared with other complexes, including mixtures of monovalent and bivalently bound antibodies, which drives the average occupancy to be greater than that one would expect for bivalent saturation. This, in turn, explains the higher observed values for the binding ratio of antigen hexamer patterns (Fig. 4c,d and Supplementary Fig. 26i). Further, modelling can accurately predict that, although about half of the hexamers are occupied by three antibodies at a high concentration (Fig. 4c,d) and settling fast at high concentrations (Fig. 4d), a majority of the structures bear other patterns of bound antibodies (Fig. 4c,d). A similar close examination of other antibody binding patterns also reveals a plethora of binding states in which the most close-packed bivalent configurations do not, in fact, constitute a majority of the states even at higher antibody concentrations (Supplementary Figs. 25–31).
Conclusions
Here we reveal, using the PSPR method to measure antibody interactions with precise nanoscale patterns of antigens, that the reach necessary for bivalent antigen binding for human IgG subclasses ranges from approximately 3 to 17 nm, with a strong peak in binding affinity at 16 nm. In addition, we show that the antibody affinity has a large effect on the spatial tolerance for bivalent binding, in effect limiting bivalent binding to the 16 nm range for low-affinity antibodies. This relationship should be generalizable to the extent that if other factors, such as configurational degrees of freedom, are held constant, then we expect a reduction in the constituent epitope affinities to reduce the functional affinity or avidity as well. Spatial tolerance thus represents a capacity to endure affinity-weakening strain on binding sites28, in addition to a reduction of that strain in the first place, for example, via flexibility. We further find that differences in the antibody constant region appear to greatly influence the binding strength to nanopatterns. IgG3 and, in particular, monomeric IgM (present on naive B cells as the BCR antigen receptor), have a much higher propensity for binding closely separated antigen pairs. This is in contrast to previous work on synthetic model systems that suggested a very modest effect of linker length and flexibility on bivalent binding strength29. In fact, our data are in line with recent studies indicating that the interaction of antibody with cognate antigen may be affected by the constant region30 and that structural differences are sufficient to cause differences in binding between isotypes of BCRs31.
Monomeric IgM BCR, anchored to the membrane of naive B cells in addition to IgD, is typically the first to encounter a pathogen. The exceptional spatial tolerance for bivalent binding of human IgM uncovered in our data (3–29 nm (Fig. 2b)) highlights an important feature of the structure of IgM. It suggests a unique ability to engage and dynamically interact with antigen patterns of a wide range of nanoscale densities, as displayed by pathogens. As B-cell responses mature in germinal centre reactions, less spatially tolerant isotypes, such as IgG, predominate. In addition to providing distinct effector functions to the antibodies, our data lead us to hypothesize that the IgG isotype switch is forcing the evolution of antibodies that bind well, despite having a lower spatial tolerance.
Further, by combining PSPR with a binding dynamics model, we dissected and characterized the distribution of antibody/antigen complexes, and thereby provide a new basis for the biophysical interrogation of these important interactions. From our study of the hexameric antigen patterns in particular, we hypothesize that the formation of hexameric IgG, which has been shown to be important for complement activation6, might actually be promoted when an antibody has a low spatial tolerance but are present at a high concentration, as opposed to an antibody with a higher spatial tolerance more likely to form bivalent attachments. The immune response progression from IgM to lower spatially tolerant antibodies, such as IgG, is consistent with such a hypothesis.
Note that the confidence and richness of the state stratification obtained from the model is due to the extensive calibration through multiple antigen patterns. Reducing this information demand should be prioritized in future work, for example, by utilizing more information-rich experimental data such as interferometric scattering microscopy32. An expanded model (Supplementary Note 3), with the incorporation of experimentally determined binding rates as a function of antigen separation distance, can predict the most prevalent binding mode to arbitrary antigen patterns. Such an analysis can provide mechanistic insights into how monoclonal antibodies engage antigens displayed on pathogens. Given a priori knowledge about the antigen pattern on the target, the antibody type involved and the strength of the antibody–antigen interaction, one could predict how the antibody will engage the target, which paves the way for more systematic studies of how antibodies with tailored antigen binding properties are able to perform effector functions to eliminate infectious agents and cancer cells.
Methods
Preparation of the p7560 and p8634 scaffold for origami
A single colony of Escherichia coli JM109 was picked and transferred to 25 ml lysogeny broth and cultured overnight at 37 °C in a shaking incubator. The culture (3 ml) was diluted into 250 ml of 2xYT and cultured in the same shaking incubator. The phages (p7560 or p8634)17 were added to the bacteria culture when its optical density reached 0.5 at a multiplicity of infection of 1, and the whole culture was incubated for an additional 5 h. The culture was transferred into a 250 ml centrifuge bottle and centrifuged twice at 4,000 relative centrifugal force (rcf) for 25 min at 4 °C, transferring into a fresh centrifuge bottle in between. PEG 8,000 (10 g) and NaCl (7.5 g) were added to the supernatant and incubated in an ice water bath for 30 min. The supernatant was then centrifuged at 10,000 rcf for 30 min at 4 °C, and in this step the phages were pelleted. The phage pellet was then resuspended in 10 ml of Tris buffer (pH 8.5), 10 ml of 0.2 M NaOH with 1% SDS was added, mixed gently by inversion and incubated for 3 min at room temperature. Afterwards, 7.5 ml of 3 M KOAc (pH 5.5) was added and the mixture was mixed gently by swirling, and incubated on ice for 10 min. The mixture was centrifuged at 16,500 rcf for 30 min at 4 °C, and the supernatant, which contained the M13 ssDNA, was added to 50 ml of 99.5% EtOH, mixed gently and incubated in an ice water bath for 30 min before centrifuging at 16,500 rcf for 30 min at 4 °C. The DNA pellet was washed with 75% EtOH, and centrifuged again at 16,500 rcf for 10 min at 4 °C. The pellet was dried at room temperature for a minimum of 15 min, and resuspended in 10 mM Tris (pH 8.5). The concentration of the ssDNA was measured with NanoDrop, and the quality and purity were characterized by agarose gel electrophoresis (1.5 % w/v agarose gel with 0.5 mg ml–1 ethidium bromide in 0.5×Tris/borate/EDTA as the running buffer).
Staple oligonucleotide preparation
Oligonucleotides were purchased from IDT in 96-well plates on a 25 nmol synthesis scale. The staples in each well were ordered to be diluted in water to a final concentration of 100 μM. The final concentration of the staples after pooling was adjusted to 400 nM each. NIP-, NP- and biotin-modified oligonucleotides were purchased from IDT.
Preparation of antigen-decorated nanostructures
Structures were designed using caDNAno33. The 18HB had been characterized previously34,35,36. The standard folding conditions used in this study were 20 nM ssDNA scaffold, 100 nM each staple oligonucleotide, 13 mM MgCl2, 5 mM Tris pH 8.5, and 1 mM EDTA. Folding was carried out by rapid heat denaturation followed by slow cooling from 80 to 60 °C over 20 min, and then from 60 to 24 °C over 14 h. Removal of excess staples was done by washing (repetitive concentration/dilution) the 18HB with PBS pH 7.4 supplemented with 10 mM MgCl2 in 100 kDa molecular weight cut-off 0.5 ml Amicon centrifugal filters (Merck Millipore).
Uncertainty estimation of distances
All double-site distances (that is, all except the ‘0’ nm) have variations that come from the linker length of two nucleotides, and from variations of the structures as a whole. The structural variations of the 18HB used for these experiments were thoroughly characterized in previous work36. From these earlier measurements, we found that the structures can vary in length due to bending and other variations with a s.d. of 3% of the total length. The linkers additionally constitute entropic springs centred around the mean distances with a maximum extension at each site of approximately 2 nm, and thus x ± 4 nm for a two-site distance, where x is the mean antigen separation. The variance of a uniform distribution with a 4 nm spread is 42/12 nm2 and the standard deviations used for the plots are calculated as the square root of the sum of these variances: σ(x) nm = √(42/12 + (0.03x)2).
Preparation of streptavidin CM3 sensor chip for SPR
A BIAcore t200 (GE Healthcare) was used to measure the binding kinetics of the antibody–antigen pairs. Streptavidin (Sigma-Aldrich) was dissolved in 100 mM sodium acetate buffer, pH 4.5, and immobilized on a CM3 chip (GE Healthcare) according to the manufacturer’s instructions. The biotinylated anchor oligonucleotide was adjusted to 200 nM in HBS-EPs running buffer, and passed through the SPR surface for 20 min, followed by a 5 min 50 mM NaOH washing.
SPR kinetic experiments
Streptavidin (10 µg ml–1 in 10 mM NaOAc, pH 4.5) was immobilized to a CM3 sensor chip (GE Healthcare) via NHS EDC (N-hydroxysuccinimide/1-ethyl-3-(3-dimethylaminopropyl) carbodiimide) coupling; the running buffer used here was 1×HBS-EP (GE Healthcare). Biotinylated anchor oligonucleotides (200 nM diluted in 1×HBS-EP running buffer) was injected over the surface for 20 min, followed by in short 5 min injection of 50 mM NaOH to wash away non-specifically bound oligonucleotides. The DNA nanostructures carrying the antigen patterns (prepared in 1×PBS supplemented with 10 mM MgCl2) were injected over the streptavidin – biotin anchor oligonucleotide surface for 20 min, followed by a 10 min buffer wash. Antibodies were diluted to various concentration ranges, depending on its affinity towards its antigen (0.025–0.5 nM for anti-DIG antibodies and 1–50 nM for antibiotin antibodies, 0.0256–1 nM for anti-NIP antibodies). A single cycle kinetics injection was used to inject the increasing concentrations of the antibodies, with each concentration having a 3 to 5 min contact time and 30 µl min–1 flow rate to avoid mass-transfer limitations, and after the highest concentration, the dissociation curved was recorded for 15 min. All antibody kinetics experiments with DNA origami patterns were carried out with 1×HBS-EP running buffer supplemented with 10 mM MgCl2. The t200 evaluation software was used to fit the data to a 1:1 Langmuir binding model and calculate the Ka, Kd and binding capacity. The surface was regenerated with a 5 min injection of 50 mM NaOH.
Transmission electron microscopy
Purified DNA nanostructures (5 μl, 10 nM) were spotted on glow-discharged, carbon-coated Formvar grids (Electron Microscopy Sciences) and incubated for 20 s before blotting with filter paper; the grid was immediately stained with 2% w/v uranyl formate solution for 20 s and finally blotting of the solution. The grids were then imaged with an FEI Morgagni 268(D) transmission electron microscope.
Gel electrophoresis
The DNA origami nanostructures were loaded (64 pmol) in 2% agarose gels that contained 0.5 mg ml–1 ethidium bromide, and the gel was run at 70 V for 3.5 h in an ice water bath. The gels were imaged with ImageQuant LAS 4000 (GE Healthcare).
Computational model
Detailed mathematical steps are described in the Supplementary Information. The model is a continuous-time Markov chain based on a chemical reaction network37,38 constructed from decomposing the antibody binding on an arbitrary 2D antigen pattern into four elementary processes (monovalent binding, monovalent unbinding, monovalent-to-bivalent conversion and bivalent-to-monovalent conversion). A breadth-first search algorithm39,40 was then executed for a given antigen pattern to discover the set of states (that is, every combination of empty antigens, monovalently occupied antigens and bivalently occupied antigens possible for a given pattern) and the transitions (each mediated by one of the four processes above) between those states. SPR kinetic profiles were generated by computing the structure’s probability to occupy a given state at each point in time beginning from an initial condition on the basis of rates (four in total, each corresponding to one of the elementary processes or parameters to be fit to the experiment) to determine the flux of state probabilities that occurs from one time step to the next41,42. Each state has a corresponding occupancy, or number of bound antibodies, associated with it that are weighted by the probability of the state to then obtain the average occupancy per structure. Experimental profiles were normalized to the number of structures, which allowed us to fit simulated binding curves to experimental curves and estimate values of the four rate parameters through minimization of the sum of squared errors. All the computations were implemented with a custom code written in Python (Supplementary Information).
Materials
All BIAcore products were purchased from GE Healthcare. All antibodies used in this study are monoclonal. Rabbit anti-DIG IgG (no. 9H27L19) was purchased from Thermo Scientific; mouse anti-DIG IgG1 (no. ab420) was purchased from Abcam; mouse antibiotin IgG1 (no. ab201341 and no. ab46862) were purchased from Abcam. Reagents for buffers and solutions (PBS, NaOH and MgCl2) were purchased from Sigma-Aldrich.
Production and purification of anti-NIP antibodies
Vectors that encode the constant heavy chains of human IgG1, IgG2, IgG3, IgG3 and IgG3-hinge engineered variants and monomeric IgM (C575S) with specificity for NIP as well as a vector that encodes the mouse λ light chain with NIP specificity were described previously43,44,45,46,47. The antibodies were produced in HEK293E cells (ATCC, CRL-1573) by transient co-transfection of the heavy- and light-chain encoding vectors using Lipofectamine 2000 (Invitrogen) following the manufacturer’s instructions or by stably transfected J558L cell lines. The antibodies were purified from collected supernatants and purified using a column coupled with NP or a CaptureSelect pre-packed anti-hIgG-CH1 column (Life Technologies) as described by the manufacturer. The collected proteins were up-concentrated and buffer changed to PBS (Sigma-Aldrich) using Amicon Ultra-15 ml 50 K columns (Millipore) prior to size exclusion chromatography using a Superdex 200 increase 10/300 GL column (GE Healthcare) coupled to an ÄKTA FPLC instrument (GE Healthcare). The monomeric fractions were up-concentrated by Amicon Ultra-0.5 ml 100K columns (Millipore). The HEK293E cell line and J558L murine myeloma cell lines were maintained in RPMI (Sigma-Aldrich,) and supplemented with 10% heat-inactivated FCS 25 µg ml–1 streptomycin and 25 U ml–1 penicillin.
Reporting Summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Code availability
All the code used for the computational results is available upon request.
Data availability
The raw data that support the plots within this paper and other findings of this study are provided in Supplementary Information and are available from the authors upon reasonable request.
Change history
19 February 2019
In the Supplementary Information file originally published with this Article, the Supplementary references 48–62 were missing; the amended file has now been uploaded.
References
Bachmann, M. F. & Jennings, G. T. Vaccine delivery: a matter of size, geometry, kinetics and molecular patterns. Nat. Rev. Immunol. 10, 787–796 (2010).
Martinez-Murillo, P. et al. Particulate array of well-ordered HIV clade C Env trimers elicits neutralizing antibodies that display a unique V2 cap approach. Immunity 46, 804–817.e7 (2017).
Kanekiyo, M. et al. Self-assembling influenza nanoparticle vaccines elicit broadly neutralizing H1N1 antibodies. Nature 499, 102–106 (2013).
Preiner, J. et al. IgGs are made for walking on bacterial and viral surfaces. Nat. Commun. 5, 4394 (2014).
Maity, P. C. et al. B cell antigen receptors of the IgM and IgD classes are clustered in different protein islands that are altered during B cell activation. Sci. Signal. 8, ra93 (2015).
Diebolder, C. A. et al. Complement is activated by IgG hexamers assembled at the cell surface. Science 343, 1260–1263 (2014).
Sondermann, P., Huber, R., Oosthuizen, V. & Jacob, U. The 3.2-Å crystal structure of the human IgG1 Fc fragment–FγRIII complex. Nature 406, 267–273 (2000).
Baker, K. et al. Neonatal Fc receptor for IgG (FcRn) regulates cross-presentation of IgG immune complexes by CD8–CD11b+ dendritic cells. Proc. Natl Acad. Sci. USA 108, 9927–9932 (2011).
Qiao, S.-W. et al. Dependence of antibody-mediated presentation of antigen on FcRn. Proc. Natl Acad. Sci. USA 105, 9337–9342 (2008).
Saphire, E. O. Crystal structure of a neutralizing human IgG against HIV-1: a template for vaccine design. Science 293, 1155–1159 (2001).
Bruhns, P. et al. Specificity and affinity of human Fc receptors and their polymorphic variants for human IgG subclasses. Blood 113, 3716–3725 (2009).
Tian, X. et al. In-depth analysis of subclass-specific conformational preferences of IgG antibodies. IUCrJ 2, 9–18 (2015).
Smith, T. J., Olson, N. H., Cheng, R. H., Chase, E. S. & Baker, T. S. Structure of a human rhinovirus-bivalently bound antibody complex: implications for viral neutralization and antibody flexibility. Proc. Natl Acad. Sci. USA 90, 7015–7018 (1993).
Zhang, X. et al. 3D structural fluctuation of IgG1 antibody revealed by individual particle electron tomography. Sci. Rep. 5, 9803 (2015).
Luedtke, R., Owen, C. S. & Karush, F. Proximity of antibody binding sites studied by fluorescence energy transfer. Biochemistry 19, 1182–1192 (1980).
Seeman, N. C. & Sleiman, H. F. DNA nanotechnology. Nat. Rev. Mater. 3, 17068 (2017).
Rothemund, P. W. K. Folding DNA to create nanoscale shapes and patterns. Nature 440, 297–302 (2006).
Douglas, S. M. et al. Self-assembly of DNA into nanoscale three-dimensional shapes. Nature 459, 414–418 (2009).
Benson, E. et al. DNA rendering of polyhedral meshes at the nanoscale. Nature 523, 441–444 (2015).
Steinhauer, C., Jungmann, R., Sobey, T. L., Simmel, F. C. & Tinnefeld, P. DNA origami as a nanoscopic ruler for superresolution microscopy. Angew. Chem. Int. Ed. 48, 8870–8873 (2009).
Nickels, P. C. et al. Molecular force spectroscopy with a DNA origami-based nanoscopic force clamp. Science 354, 305–307 (2016).
Fu, J. et al. Multi-enzyme complexes on DNA scaffolds capable of substrate channelling with an artificial swinging arm. Nat. Nanotech. 9, 531–536 (2014).
Wollman, A. J. M., Sanchez-Cano, C., Carstairs, H. M. J., Cross, R. A. & Turberfield, A. J. Transport and self-organization across different length scales powered by motor proteins and programmed by DNA. Nat. Nanotech. 9, 44–47 (2013).
Funke, J. J. & Dietz, H. Placing molecules with Bohr radius resolution using DNA origami. Nat. Nanotech. 11, 47–52 (2015).
Rinker, S., Ke, Y., Liu, Y., Chhabra, R. & Yan, H. Self-assembled DNA nanostructures for distance-dependent multivalent ligand–protein binding. Nat. Nanotech. 3, 418–422 (2008).
McKinney, S. A., Joo, C. & Ha, T. Analysis of single-molecule FRET trajectories using hidden Markov modeling. Biophys. J. 91, 1941–1951 (2006).
Müller, K. M., Arndt, K. M. & Plückthun, A. Model and simulation of multivalent binding to fixed ligands. Anal. Biochem. 261, 149–158 (1998).
Natkanski, E. et al. B cells use mechanical energy to discriminate antigen affinities. Science 340, 1587–1590 (2013).
Mack, E. T., Snyder, P. W., Perez-Castillejos, R. & Whitesides, G. M. Using covalent dimers of human carbonic anhydrase II to model bivalency in immunoglobulins. J. Am. Chem. Soc. 133, 11701–11715 (2011).
Sela-Culang, I., Kunik, V. & Ofran, Y. The structural basis of antibody–antigen recognition. Front. Immunol. 4, 1–13 (2013).
Übelhart, R. et al. Responsiveness of B cells is regulated by the hinge region of IgD. Nat. Immunol. 16, 534–543 (2015).
Cole, D., Young, G., Weigel, A., Sebesta, A. & Kukura, P. Label-free single-molecule imaging with numerical-aperture-shaped interferometric scattering microscopy. ACS Photon. 4, 211–216 (2017).
Douglas, S. M. et al. Rapid prototyping of 3D DNA-origami shapes with caDNAno. Nucleic Acids Res. 37, 5001–5006 (2009).
Shaw, A. et al. Spatial control of membrane receptor function using ligand nanocalipers. Nat. Methods 11, 841–846 (2014).
Zhao, Y.-X. et al. DNA origami delivery system for cancer therapy with tunable release properties. ACS Nano 6, 8684–8691 (2012).
Reuss, M. et al. Measuring true localization accuracy in super resolution microscopy with DNA-origami nanostructures. New J. Phys. 19, 025013 (2017).
Gillespie, D. T. Exact stochastic simulation of coupled chemical reactions. J. Phys. Chem. 81, 2340–2361 (1977).
Kurtz, T. G. The relationship between stochastic and deterministic models for chemical reactions. J. Chem. Phys. 57, 2976–2978 (1972).
Moore, E. F. in Proceedings of the International Symposium on the Theory of Switching 285–292 (Harvard Univ. Press, 1959).
Lee, C. Y. An algorithm for path connections and its applications. IRE Trans. Electron. Comput. EC 10, 346–365 (1961).
Fox, B. L. & Glynn, P. W. Computing Poisson probabilities. Commun. ACM 31, 440–445 (1988).
Gross, D. & Miller, D. R. The randomization technique as a modeling tool and solution procedure for transient Markov processes. Oper. Res. 32, 343–361 (1984).
Michaelsen, T. E., Garred, P. & Aase, A. Human IgG subclass pattern of inducing complement-mediated cytolysis depends on antigen concentration and to a lesser extent on epitope patchiness, antibody affinity and complement concentration. Eur. J. Immunol. 21, 11–16 (1991).
Norderhaug, L., Olafsen, T., Michaelsen, T. E. & Sandlie, I. Versatile vectors for transient and stable expression of recombinant antibody molecules in mammalian cells. J. Immunol. Methods 204, 77–87 (1997).
Berntzen, G. et al. Prolonged and increased expression of soluble Fc receptors, IgG and a TCR-Ig fusion protein by transiently transfected adherent 293E cells. J. Immunol. Methods 298, 93–104 (2005).
Michaelsen, T. E. et al. One disulfide bond in front of the second heavy chain constant region is necessary and sufficient for effector functions of human IgG3 without a genetic hinge. Proc. Natl Acad. Sci. USA 91, 9243–9247 (1994).
Brekke, O. H., Michaelsen, T. E., Sandin, R. & Sandlie, I. Activation of complement by an IgG molecule without a genetic hinge. Nature 363, 628–630 (1993).
Acknowledgements
This work was funded through grants from the Swedish Research Council (grant no. 2013–5883 to B.H.), the Swedish Foundation for Strategic Research (grant FFL12–0219 to B.H.), the European Research Council (ERC) under the European Union’s Horizon 2020 research and innovation programme (grant agreement no. 724872 to B.H.) and the Knut and Alice Wallenberg foundation (Academy Fellow grant KAW2014.0241 to B.H.). The SPR instrument was funded by the SFO initiative StatRegen at Karolinska Institutet. J.T.A. was in part supported by the Research Council of Norway through its Centres of Excellence funding scheme (project no. 179573) and the Research Council of Norway (grant no. 230526/F20 and no. 274993/O70). We thank S. Foss for help with schematic antibody figures. We thank Y. Bryceson and S. Chiang for work on the experimental testing and discussions, and G. Karlsson Hedestam and A. Teixeira for discussions.
Author information
Authors and Affiliations
Contributions
A.S., J.T.A. and B.H. conceived the study and wrote the paper. J.T.A., I.Sa., A.G. and T.E.M. helped conceive the human antibody panel study. A.S. performed most of the experimental work on origami and SPR. I.T.H. performed most of the theoretical and modelling work. I.Sm. performed a significant proportion of the experimental work on origami and SPR. J.R. performed origami experiments and design. J.T.A., D.B., T.E.M. and A.G. performed the production and purification of the human antibody variants. All the authors contributed to writing the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary notes 1–4, Supplementary Figures 1–31, Supplementary Tables 1–5
Rights and permissions
About this article
Cite this article
Shaw, A., Hoffecker, I.T., Smyrlaki, I. et al. Binding to nanopatterned antigens is dominated by the spatial tolerance of antibodies. Nature Nanotech 14, 184–190 (2019). https://doi.org/10.1038/s41565-018-0336-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41565-018-0336-3
This article is cited by
-
Super-resolved FRET and co-tracking in pMINFLUX
Nature Photonics (2024)
-
Enhancing antibody responses by multivalent antigen display on thymus-independent DNA origami scaffolds
Nature Communications (2024)
-
A DNA origami device spatially controls CD95 signalling to induce immune tolerance in rheumatoid arthritis
Nature Materials (2024)
-
Digital nanoreactors to control absolute stoichiometry and spatiotemporal behavior of DNA receptors within lipid bilayers
Nature Communications (2023)
-
Commercial influenza vaccines vary in HA-complex structure and in induction of cross-reactive HA antibodies
Nature Communications (2023)