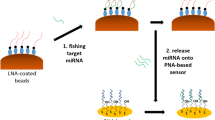
Abstract
There is intense interest in quantifying the levels of microRNA because of its importance as a blood-borne biomarker. The challenge has been to develop methods that can monitor microRNA expression both over broad concentration ranges and in ultralow amounts directly in a patient’s blood. Here, we show that, through electric-field-induced reconfiguration of a network of gold-coated magnetic nanoparticles modified by probe DNA (DNA–Au@MNPs), it is possible to create a highly sensitive sensor for direct analysis of nucleic acids in samples as complex as whole blood. The sensor is the first to be able to detect concentrations of microRNA from 10 aM to 1 nM in unprocessed blood samples. It can distinguish small variations in microRNA concentrations in blood samples of mice with growing tumours. The ultrasensitive and direct detection of microRNA using an electrically reconfigurable DNA–Au@MNPs network makes the reported device a promising tool for cancer diagnostics.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Alhasan, A. H. et al. Circulating microRNA signature for the diagnosis of very high-risk prostate cancer. Proc. Natl Acad. Sci. USA 113, 10655–10660 (2016).
Mohammadi, M., Goodarzi, M., Jaafari, M. R., Mirzaei, H. R. & Mirzaei, H. Circulating microRNA: a new candidate for diagnostic biomarker in neuroblastoma. Cancer Gene Ther. 23, 371–372 (2016).
Nadal, E. et al. A novel serum 4-microRNA signature for lung cancer detection. Sci. Rep. 5, 12464 (2015).
Schwarzenbach, H., Nishida, N., Calin, G. A. & Pantel, K. Clinical relevance of circulating cell-free microRNAs in cancer. Nat. Rev. Clin. Oncol. 11, 145–156 (2014).
Wozniak, M. B. et al. Circulating microRNAs as non-invasive biomarkers for early detection of non-small-cell lung cancer. PLoS ONE 10, e0125026 (2015).
El-Khoury, V., Pierson, S., Kaoma, T., Bernardin, F. & Berchem, G. Assessing cellular and circulating miRNA recovery: the impact of the RNA isolation method and the quantity of input material. Sci. Rep. 6, 19529 (2016).
Pritchard, C. C., Cheng, H. H. & Tewari, M. MicroRNA profiling: approaches and considerations. Nat. Rev. Genet. 13, 358–369 (2012).
Haun, J. B., Yoon, T. J., Lee, H. & Weissleder, R. Magnetic nanoparticle biosensors. Wiley Interdisc. Rev. Nanomed. Nanobiotechnol. 2, 291–304 (2010).
Farina, N. H. et al. Standardizing analysis of circulating microRNA: clinical and biological relevance. J. Cell. Biochem. 115, 805–811 (2014).
Labib, M., Sargent, E. H. & Kelley, S. O. Electrochemical methods for the analysis of clinically relevant biomolecules. Chem. Rev. 116, 9001–9090 (2016).
Tavallaie, R., De Almeida, S. R. & Gooding, J. J. Toward biosensors for the detection of circulating microRNA as a cancer biomarker: an overview of the challenges and successes. Wiley Interdisc. Rev. Nanomed. Nanobiotechnol. 7, 580–592 (2015).
Tavallaie, R., Darwish, N., Gebala, M., Hibbert, D. B. & Gooding, J. J. The effect of interfacial design on the electrochemical detection of DNA and microRNA using methylene blue at low-density DNA films. ChemElectroChem 1, 165–171 (2014).
Gooding, J. J. & Gaus, K. Single-molecule sensors: challenges and opportunities for quantitative analysis. Angew. Chem. Int. Ed. 55, 11354–11366 (2016).
Hsieh, K., Ferguson, B. S., Eisenstein, M., Plaxco, K. W. & Soh, H. T. Integrated electrochemical microsystems for genetic detection of pathogens at the point of care. Acc. Chem. Res. 48, 911–920 (2015).
Cardoso, A. R., Moreira, F. T. C., Fernandes, R. & Sales, M. G. F. Novel and simple electrochemical biosensor monitoring attomolar levels of miRNA-155 in breast cancer. Biosens. Bioelectron. 80, 621–630 (2016).
Miao, P., Wang, B. D., Meng, F. Y., Yin, J. & Tang, Y. G. Ultrasensitive detection of microRNA through rolling circle amplification on a DNA tetrahedron decorated electrode. Bioconjug. Chem. 26, 602–607 (2015).
Ramnani, P., Gao, Y. N., Ozsoz, M. & Mulchandani, A. Electronic detection of microRNA at attomolar level with high specificity. Anal. Chem. 85, 8061–8064 (2013).
Soleymani, L., Fang, Z. C., Sargent, E. H. & Kelley, S. O. Programming the detection limits of biosensors through controlled nanostructuring. Nat. Nanotech. 4, 844–848 (2009).
Wang, Y., Zheng, D. L., Tan, Q. L., Wang, M. X. & Gu, L. Q. Nanopore-based detection of circulating microRNAs in lung cancer patients. Nat. Nanotech. 6, 668–674 (2011).
Bakshi, S. F. et al. Magnetic field-activated sensing of mRNA in living cells. J. Am. Chem. Soc. 139, 12117–12120 (2017).
Goon, I. Y., Lai, L. M. H., Lim, M., Amal, R. & Gooding, J. J. ‘Dispersible electrodes’: a solution to slow response times of sensitive sensors. Chem. Commun. 46, 8821–8823 (2010).
Lai, L. M. H. et al. Gold-coated magnetic nanoparticles as ‘dispersible electrodes’ – understanding their electrochemical performance. J. Electroanal. Chem. 656, 130–135 (2011).
Chuah, K. et al. Ultrasensitive electrochemical detection of prostate-specific antigen (PSA) using gold-coated magnetic nanoparticles as ‘dispersible electrodes’. Chem. Commun. 48, 3503–3505 (2012).
Markou, A., Zavridou, M. & Lianidou, E. S. miRNA-21 as a novel therapeutic target in lung cancer. Lung Cancer 7, 19–27 (2016).
Goon, I. Y. et al. Fabrication and dispersion of gold-shell-protected magnetite nanoparticles: systematic control using polyethyleneimine. Chem. Mater. 21, 673–681 (2009).
Selcuklu, S. D., Donoghue, M. T. A. & Spillane, C. miR-21 as a key regulator of oncogenic processes. Biochem. Soc. Trans. 37, 918–925 (2009).
Yang, Y. et al. Downregulation of microRNA-21 expression restrains non-small cell lung cancer cell proliferation and migration through upregulation of programmed cell death 4. Cancer Gene Ther. 22, 23–29 (2015).
Tavallaie, R., Darwish, N., Hibbert, D. B. & Gooding, J. J. Nucleic-acid recognition interfaces: how the greater ability of RNA duplexes to bend towards the surface influences electrochemical sensor performance. Chem. Commun. 51, 16526–16529 (2015).
Lai, L. M. H. et al. The biochemiresistor: an ultrasensitive biosensor for small organic molecules. Angew. Chem. Int. Ed, 51, 6456–6459 (2012).
Dejima, H., Iinuma, H., Kanaoka, R., Matsutani, N. & Kawamura, M. Exosomal microRNA in plasma as a non-invasive biomarker for the recurrence of non-small cell lung cancer. Oncol. Lett. 13, 1256–1263 (2017).
Fischer, L. M. et al. Gold cleaning methods for electrochemical detection applications. Microelectron. Eng. 86, 1282–1285 (2009).
McCarroll, J. A. et al. TUBB3//βIII-tubulin acts through the PTEN/AKT signaling axis to promote tumorigenesis and anoikis resistance in non-small cell lung cancer. Cancer Res. 75, 415–425 (2015).
Acknowledgements
The authors acknowledge support from the UNSW Mark Wainwright Analytical Centre, Biological Resources Imaging Laboratory and Electron Microscope Unit. The authors also acknowledge assistance from K. Kimpton with in vivo mouse experiments. The authors acknowledge funding from the ARC Centre of Excellence in Convergent Bio-Nano Science and Technology (J.J.G. and M.K.; CE140100036), the ARC Laureate Fellowship (J.J.G.; FL150100060) programme, a National Health and Medical Research Council programme grant (M.K. and J.J.G.; APP1091261) and an NHMRC Principal Research Fellowship (M.K.; APP1119152). J.M. is supported by a Cancer Institute NSW Career Development Fellowship.
Author information
Authors and Affiliations
Contributions
R.T. and J.J.G. designed the experiments, performed data interpretation and wrote the manuscript. R.T. performed the experiments and analysed the data. J.M. performed miR-21 inhibitor transfections and in vivo experiments, with assistance from M.L.G. N.A. and R.D.T. performed the electron microscopy. J.M. and M.K. provided scientific and technical support and data interpretation in biological experiments. W.S. and E.B. provided scientific support in the creation and investigation of the hypothesis for the mechanism of detection. D.B.H. provided scientific support in performing calculations required for the design of experiments. All authors reviewed the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary Figures 1–7 and Supplementary References
Rights and permissions
About this article
Cite this article
Tavallaie, R., McCarroll, J., Le Grand, M. et al. Nucleic acid hybridization on an electrically reconfigurable network of gold-coated magnetic nanoparticles enables microRNA detection in blood. Nature Nanotech 13, 1066–1071 (2018). https://doi.org/10.1038/s41565-018-0232-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41565-018-0232-x
This article is cited by
-
Versatile magnetic configuration for the control and manipulation of superparamagnetic nanoparticles
Scientific Reports (2023)
-
Analytical device miniaturization for the detection of circulating biomarkers
Nature Reviews Bioengineering (2023)
-
Multiplexed RNA profiling by regenerative catalysis enables blood-based subtyping of brain tumors
Nature Communications (2023)
-
Affinity-based electrochemical sensors for biomolecular detection in whole blood
Analytical and Bioanalytical Chemistry (2023)
-
Localized DNA tetrahedrons assisted catalytic hairpin assembly for the rapid and sensitive profiling of small extracellular vesicle-associated microRNAs
Journal of Nanobiotechnology (2022)