Abstract
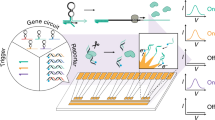
In synthetic biology, the control of gene expression requires a multistep processing of biological signals. The key steps are sensing the environment, computing information and outputting products1. To achieve such functions, the laborious, combinational networking of enzymes and substrate-genes is required, and to resolve problems, sophisticated design automation tools have been introduced2. However, the complexity of genetic circuits remains low because it is difficult to completely avoid crosstalk between the circuits. Here, we have made an orthogonal self-contained device by integrating an actuator and sensors onto a DNA origami-based nanochip that contains an enzyme, T7 RNA polymerase (RNAP) and multiple target-gene substrates. This gene nanochip orthogonally transcribes its own genes, and the nano-layout ability of DNA origami allows us to rationally design gene expression levels by controlling the intermolecular distances between the enzyme and the target genes. We further integrated reprogrammable logic gates so that the nanochip responds to water-in-oil droplets and computes their small RNA (miRNA) profiles, which demonstrates that the nanochip can function as a gene logic-chip. Our approach to component integration on a nanochip may provide a basis for large-scale, integrated genetic circuits.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Gardner, T. S., Cantor, C. R. & Collins, J. J. Construction of a genetic toggle switch in Escherichia coli. Nature 403, 339–342 (2000).
Nielsen, A. A. et al. Genetic circuit design automation. Science 352, aac7341 (2016).
Maniatis, T. & Reed, R. An extensive network of coupling among gene expression machines. Nature 416, 499–506 (2002).
Rothemund, P. W. K. Folding DNA to create nanoscale shapes and patterns. Nature 440, 297–302 (2006).
Douglas, S. M. et al. Self-assembly of DNA into nanoscale three-dimensional shapes. Nature 459, 414–418 (2009).
Iinuma, R. et al. Polyhedra self-assembled from DNA tripods and characterized with 3D DNA-PAINT. Science 344, 65–69 (2014).
Seelig, G., Soloveichik, D., Zhang, D. Y. & Winfree, E. Enzyme-free nucleic acid logic circuits. Science 314, 1585–1588 (2006).
Douglas, S. M., Bachelet, I. & Church, G. M. A logic-gated nanorobot for targeted transport of molecular payloads. Science 335, 831–834 (2012).
Simmel, F. C. DNA-based assembly lines and nanofactories. Curr. Opin. Biotechnol. 23, 516–521 (2012).
Delebecque, C. J., Lindner, A. B., Silver, P. A. & Aldaye, F. A. Organization of intracellular reactions with rationally designed RNA assemblies. Science 333, 470–474 (2011).
Chatterjee, G., Dalchau, N., Muscat, R. A., Phillips, A. & Seelig, G. A spatially localized architecture for fast and modular DNA computing. Nat. Nanotech. 12, 920–927 (2017).
Wilner, O. I. et al. Enzyme cascades activated on topologically programmed DNA scaffolds. Nat. Nanotech. 4, 249–254 (2009).
Fu, J., Liu, M., Liu, Y., Woodbury, N. W. & Yan, H. Interenzyme substrate diffusion for an enzyme cascade organized on spatially addressable DNA nanostructures. J. Am. Chem. Soc. 134, 5516–5519 (2012).
Fu, J. et al. Multi-enzyme complexes on DNA scaffolds capable of substrate channelling with an artificial swinging arm. Nat. Nanotech. 9, 531–536 (2014).
Zhao, Z. et al. Nanocaged enzymes with enhanced catalytic activity and increased stability against protease digestion. Nat. Commun. 7, 10619 (2016).
Wickham, S. F. J. et al. Direct observation of stepwise movement of a synthetic molecular transporter. Nat. Nanotech. 6, 166–169 (2011).
Sacca, B. et al. Orthogonal protein decoration of DNA origami. Angew. Chem. Int. Ed. 49, 9378–9383 (2010).
Zhang, Y., Tsitkov, S. & Hess, H. Proximity does not contribute to activity enhancement in the glucose oxidase-horseradish peroxidase cascade. Nat. Commun. 7, 13982 (2016).
Bustamante, C., Marko, J. F., Siggia, E. D. & Smith, S. Entropic elasticity of lambda-phage DNA. Science 265, 1599–1600 (1994).
Miyazono, Y., Hayashi, M., Karagiannis, P., Harada, Y. & Tadakuma, H. Strain through the neck linker ensures processive runs: a DNA–kinesin hybrid nanomachine study. EMBO J. 29, 93–106 (2010).
Shimizu, Y. et al. Cell-free translation reconstituted with purified components. Nat. Biotechnol. 19, 751–755 (2001).
Nakamura, K. et al. Culture-independent method for identification of microbial enzyme-encoding genes by activity-based single-cell sequencing using a water-in-oil microdroplet platform. Sci. Rep. 6, 22259 (2016).
Yao, C., Sasaki, H. M., Ueda, T., Tomari, Y. & Tadakuma, H. Single-molecule analysis of the target cleavage reaction by the Drosophila RNAi enzyme complex. Mol. Cell 59, 125–132 (2015).
Yoshimura, Y. & Fujimoto, K. Ultrafast reversible photo-cross-linking reaction: toward in situ DNA manipulation. Org. Lett. 10, 3227–3230 (2008).
Zhou, Z. P. et al. Single molecule imaging of the trans-translation entry process via anchoring of the tagged ribosome. J. Biochem. 149, 609–618 (2010).
Jinek, M. et al. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 337, 816–821 (2012).
Praetorius, F. & Dietz, H. Self-assembly of genetically encoded DNA-protein hybrid nanoscale shapes. Science 355, eaam5488 (2017).
Rinker, S., Ke, Y., Liu, Y., Chhabra, R. & Yan, H. Self-assembled DNA nanostructures for distance-dependent multivalent ligand-protein binding. Nat. Nanotech. 3, 418–422 (2008).
Williams, B. A., Lund, K., Liu, Y., Yan, H. & Chaput, J. C. Self-assembled peptide nanoarrays: an approach to studying protein–protein interactions. Angew. Chem. Int. Ed. 46, 3051–3054 (2007).
Bonnet, J., Yin, P., Ortiz, M. E., Subsoontorn, P. & Endy, D. Amplifying genetic logic gates. Science 340, 599–603 (2013).
Bentele, K., Saffert, P., Rauscher, R., Ignatova, Z. & Bluthgen, N. Efficient translation initiation dictates codon usage at gene start. Mol. Syst. Biol. 9, 675 (2013).
Derr, N. D. et al. Tug-of-war in motor protein ensembles revealed with a programmable DNA origami scaffold. Science 338, 662–665 (2012).
Torisawa, T. et al. Autoinhibition and cooperative activation mechanisms of cytoplasmic dynein. Nat. Cell Biol. 16, 1118–1124 (2014).
Haneoka, M. et al. Microfluidic active sorting of DNA molecules labeled with single quantum dots using flow switching by a hydrogel sol–gel transition. Sens. Actuat. B 159, 314–320 (2011).
Qin, D., Xia, Y. & Whitesides, G. M. Soft lithography for micro- and nanoscale patterning. Nat. Protoc. 5, 491–502 (2010).
Okano, T., Matsuura, T., Kazuta, Y., Suzuki, H. & Yomo, T. Cell-free protein synthesis from a single copy of DNA in a glass microchamber. Lab Chip 12, 2704–2711 (2012).
Thirumalai, D. & Ha B. Y. in Theoretical and Mathematical Models in Polymer Research (ed. Grosberg, A.) 1–35 (Academia, New York, 1988).
Schuler, B., Lipman, E. A., Steinbach, P. J., Kumke, M. & Eaton, W. A. Polyproline and the ‘spectroscopic ruler’ revisited with single-molecule fluorescence. Proc. Natl Acad. Sci. USA 102, 2754–2759 (2005).
Kitano, H., Funahashi, A., Matsuoka, Y. & Oda, K. Using process diagrams for the graphical representation of biological networks. Nat. Biotechnol. 23, 961–966 (2005).
Dörr, A., Keller, R., Zell, A. & Drager, A. SBMLsimulator: a Java tool for model simulation and parameter estimation in systems biology. Computation 2, 246–257 (2014).
Acknowledgements
The authors thank RIBM for AFM imaging, T. Sakurai, H. Okada and K. Nakamura (The University of Tokyo) for help with construction of the microfluidics system, and Y. Tomari and members of the Tomari Laboratory for providing comments on the miRNA studies and on this manuscript. The authors also thank Y. Hatano (Osaka University) and M. Hagiya (The University of Tokyo) for their comments on logic circuits, and I. Kawamata (Tohoku University) for comments on the kinetically based simulation of logic circuit. This work was partially supported by Grants-in-Aid for Scientific Research on Innovative Areas (‘Molecular Robotics’ to H.T. and M.E., nos. 15H00798 and 24104002; ‘Non-coding RNA Neo-taxonomy’ to H.T., no. 26113007), a Grant-in-Aid for Young Scientists (A), a Grant-in-Aid for Scientific Research (B) and a Grant-in-Aid for Challenging Exploratory Research (to H.T., nos. 24687018, 16KT0068 and 15K14485), Grants-in-Aid for Scientific Research (S) (to Y.H., no. 26220602) and (A) (to S.S., no. 16H02349), Grants-in-Aid for Young Scientists (B) (to R.I., no. 15K18668) from the Japan Ministry of Education, Culture, Sports, Science and Technology, Research Fellowships for Young Scientists (to T.M., no. 15J08491), Core-to-Core Program, A, Advanced Research Networks (Phototheranostics) (to R.I.) from the Japan Society for the Promotion of Science, CREST (to Y.H., no. JPMJCR1333), the Centre of Innovation (COI) Program (to T.F.) from the Japan Science and Technology Agency, the Cooperative Research Program of the Institute for Protein Research, Osaka University (CRa-18-01, to H.T and M.E.), the Asahi Glass Foundation, Futaba Electronics Memorial Foundation, and Hamaguchi Foundation for the Advancement of Biochemistry (to H.T.), and Futaba Electronics Memorial Foundation Scholarship (to T.M.).
Author information
Authors and Affiliations
Contributions
H.T. conceived, designed and supervised the study. T.M. performed the biochemical and microfluidics experiments and analysed the data. H.T. performed the AFM imaging with the help of RIBM (Tsukuba, Japan). R.I., T.F., S.S., A.I., D.H.Y, and T.S. constructed the microfluidics system. T.U. supervised H.T. and T.M. All authors discussed the results. H.T., H.S., T.M. and M.E. wrote the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors have a pending patent application on the programmable gene expression method in this work.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary Figures 1–30, Supplementary Tables 1–4, Supplementary Notes and Supplementary References
Rights and permissions
About this article
Cite this article
Masubuchi, T., Endo, M., Iizuka, R. et al. Construction of integrated gene logic-chip. Nature Nanotech 13, 933–940 (2018). https://doi.org/10.1038/s41565-018-0202-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41565-018-0202-3
This article is cited by
-
A genetic circuit on a single DNA molecule as an autonomous dissipative nanodevice
Nature Communications (2024)
-
Fabricating higher-order functional DNA origami structures to reveal biological processes at multiple scales
NPG Asia Materials (2023)
-
Three-dimensional electron ptychography of organic–inorganic hybrid nanostructures
Nature Communications (2022)
-
Functionalizing DNA origami to investigate and interact with biological systems
Nature Reviews Materials (2022)
-
Proximity-induced caspase-9 activation on a DNA origami-based synthetic apoptosome
Nature Catalysis (2020)