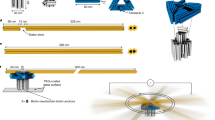
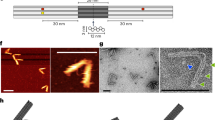
Abstract
Dynamic DNA nanotechnology has yielded nontrivial autonomous behaviours such as stimulus-guided locomotion, computation and programmable molecular assembly. Despite these successes, DNA-based nanomachines suffer from slow kinetics, requiring several minutes or longer to carry out a handful of operations. Here, we pursue the speed limit of an important class of reactions in DNA nanotechnology—toehold exchange—through the single-molecule optimization of a novel class of DNA walker that undergoes cartwheeling movements over a field of complementary oligonucleotides. After optimizing this DNA ‘acrobat’ for rapid movement, we measure a stepping rate constant approaching 1 s−1, which is 10- to 100-fold faster than prior DNA walkers. Finally, we use single-particle tracking to demonstrate movement of the walker over hundreds of nanometres within 10 min, in quantitative agreement with predictions from stepping kinetics. These results suggest that substantial improvements in the operating rates of broad classes of DNA nanomachines utilizing strand displacement are possible.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Sherman, W. B. & Seeman, N. C. A precisely controlled DNA biped walking device. Nano Lett. 4, 1203–1207 (2004).
Shin, J.-S. & Pierce, N. A. A synthetic DNA walker for molecular transport. J. Am. Chem. Soc. 126, 10834–10835 (2004).
Tian, Y., He, Y., Chen, Y., Yin, P. & Mao, C. A DNAzyme that walks processively and autonomously along a one-dimensional track. Angew. Chem. Int. Ed. Engl. 44, 4355–4358 (2005).
Bath, J., Green, S. J. & Turberfield, A. J. A free-running DNA motor powered by a nicking enzyme. Angew. Chem. Int. Ed. Engl. 44, 4358–4361 (2005).
Omabegho, T., Sha, R. & Seeman, N. C. A bipedal DNA Brownian motor with coordinated legs. Science 324, 67–71 (2009).
Gu, H., Chao, J., Xiao, S.-J. & Seeman, N. C. A proximity-based programmable DNA nanoscale assembly line. Nature 465, 202–205 (2010).
He, Y. & Liu, D. R. Autonomous multistep organic synthesis in a single isothermal solution mediated by a DNA walker. Nat. Nanotech. 5, 778–782 (2010).
Qian, L. & Winfree, E. Scaling up digital circuit computation with DNA strand displacement cascades. Science 332, 1196–1201 (2011).
Qian, L., Winfree, E. & Bruck, J. Neural network computation with DNA strand displacement cascades. Nature 475, 368–372 (2011).
Lund, K. et al. Molecular robots guided by prescriptive landscapes. Nature 465, 206–210 (2010).
Douglas, S. M., Bachelet, I. & Church, G. M. A logic-gated nanorobot for targeted transport of molecular payloads. Science 335, 831–834 (2012).
Thubagere, A. J. et al. A cargo-sorting DNA robot. Science 357, eaan6558 (2017).
Zhang, D. Y. & Seelig, G. Dynamic DNA nanotechnology using strand-displacement reactions. Nat. Chem. 3, 103–113 (2011).
Zhang, D. Y. & Winfree, E. Control of DNA strand displacement kinetics using toehold exchange. J. Am. Chem. Soc. 131, 17303–17314 (2009).
Zhang, D. Y., Chen, S. X. & Yin, P. Optimizing the specificity of nucleic acid hybridization. Nat. Chem. 4, 208–214 (2012).
Fu, J. et al. Multi-enzyme complexes on DNA scaffolds capable of substrate channelling with an artificial swinging arm. Nat. Nanotech. 9, 531–536 (2014).
Wickham, S. F. J. et al. Direct observation of stepwise movement of a synthetic molecular transporter. Nat. Nanotech. 6, 166–169 (2011).
Cha, T.-G. et al. A synthetic DNA motor that transports nanoparticles along carbon nanotubes. Nat. Nanotech. 9, 39–43 (2014).
Yehl, K. et al. High-speed DNA-based rolling motors powered by RNase H. Nat. Nanotech. 11, 184–190 (2015).
King, S. J. & Schroer, T. A. Dynactin increases the processivity of the cytoplasmic dynein motor. Nat. Cell Biol. 2, 20–24 (2000).
Kural, C. et al. Kinesin and dynein move a peroxisome in vivo: a tug-of-war or coordinated movement? Science 308, 1469–1472 (2005).
Pan, J., Li, F., Cha, T.-G., Chen, H. & Choi, J. H. Recent progress on DNA based walkers. Curr. Opin. Biotechnol. 34, 56–64 (2015).
Roy, R., Hohng, S. & Ha, T. A practical guide to single-molecule FRET. Nat. Methods 5, 507–516 (2008).
Walter, N. G., Huang, C.-Y., Manzo, A. J. & Sobhy, M. A. Do-it-yourself guide: how to use the modern single-molecule toolkit. Nat. Methods 5, 475–489 (2008).
Srinivas, N. et al. On the biophysics and kinetics of toehold-mediated DNA strand displacement. Nucleic Acids Res. 41, 10641–10658 (2013).
Panyutin, I. G. & Hsieh, P. Formation of a single base mismatch impedes spontaneous DNA branch migration. J. Mol. Biol. 230, 413–424 (1993).
Beattie, K. L., Wiegand, R. C. & Radding, C. M. Uptake of homologous single-stranded fragments by superhelical DNA. J. Mol. Biol. 116, 783–803 (1977).
Jungmann, R. et al. Single-molecule kinetics and super-resolution microscopy by fluorescence imaging of transient binding on DNA origami. Nano Lett. 10, 4756–4761 (2010).
Dupuis, N. F., Holmstrom, E. D. & Nesbitt, D. J. Single-molecule kinetics reveal cation-promoted DNA duplex formation through ordering of single-stranded helices. Biophys. J. 105, 756–766 (2013).
Smith, S. B., Cui, Y. & Bustamante, C. Overstretching B-DNA: the elastic response of individual double-stranded and single-stranded DNA molecules. Science 271, 795–799 (1996).
Helenius, J., Brouhard, G., Kalaidzidis, Y., Diez, S. & Howard, J. The depolymerizing kinesin MCAK uses lattice diffusion to rapidly target microtubule ends. Nature 441, 115–119 (2006).
Ha, T. Single-molecule fluorescence resonance energy transfer. Methods 25, 78–86 (2001).
Michelotti, N., de Silva, C., Johnson-Buck, A. E., Manzo, A. J. & Walter, N. G. A bird’s eye view tracking slow nanometer-scale movements of single molecular nano-assemblies. Methods Enzymol. 475, 121–148 (2010).
Aitken, C. E., Marshall, R. A. & Puglisi, J. D. An oxygen scavenging system for improvement of dye stability in single-molecule fluorescence experiments. Biophys. J. 94, 1826–1835 (2008).
Blanco, M. & Walter, N. G. Analysis of complex single-molecule FRET time trajectories. Methods Enzymol. 472, 153–178 (2010).
Nicolai, C. & Sachs, F. Solving ion channel kinetics with the qub software. Biophys. Rev. Lett. 08, 191–211 (2013).
Dill, K. A. & Bromberg, S. Molecular Driving Forces: Statistical Thermodynamics in Chemistry and Biology. (Garland Science: New York, 2003).
Chen, H. et al. Ionic strength-dependent persistence lengths of single-stranded RNA and DNA. Proc. Natl Acad. Sci. USA 109, 799–804 (2012).
Johnson-Buck, A. et al. Kinetic fingerprinting to identify and count single nucleic acids. Nat. Biotechnol. 33, 730–732 (2015).
Particle Track and Analysis (PTA) (Yoshiyuki Arai, accessed 29 August 2017); http://www.sanken.osaka-u.ac.jp/labs/bse/ImageJcontents/frameImageJ.html
Gillespie, D. T. Exact stochastic simulation of coupled chemical reactions. J. Phys. Chem. 81, 2340–2361 (1977).
Acknowledgements
This work was supported primarily by the US Department of Defense Army Research Office MURI award W911NF-12-1-0420 to N.G.W. and H.Y. The authors thank J. Damon Hoff for technical support, Z. R. Li for graphic design support, and B. Nijholt, E. Krieg, A. M. Bergman and W. Benjamin Rogers for discussions about DNA origami design.
Author information
Authors and Affiliations
Contributions
A.J.-B. conceived the ideas. Y.R.Y. designed, fabricated and characterized the DNA tile samples. W.M.S. and A.J.-B. designed, fabricated and characterized the DNA origami samples. J.L. and A.J.-B. performed smFRET and single-particle tracking measurements. J.L., A.J.-B., and N.G.W. analysed and interpreted the data. J.L. and A.J.-B. co-wrote the paper, and all authors discussed the results and edited the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary Text, Supplementary Figures 1–34, Supplementary Tables 1–5 and Supplementary References
Rights and permissions
About this article
Cite this article
Li, J., Johnson-Buck, A., Yang, Y.R. et al. Exploring the speed limit of toehold exchange with a cartwheeling DNA acrobat. Nature Nanotech 13, 723–729 (2018). https://doi.org/10.1038/s41565-018-0130-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41565-018-0130-2
This article is cited by
-
Bispecific aptamer-initiated 3D DNA nanomotor biosensor powered by DNAzyme and entropy-driven circuit for sensitive and specificity detection of lysozyme
Nano Research (2023)
-
Nucleic acid-based scaffold systems and application in enzyme cascade catalysis
Applied Microbiology and Biotechnology (2023)
-
A synthetic tubular molecular transport system
Nature Communications (2021)
-
Automatic classification and segmentation of single-molecule fluorescence time traces with deep learning
Nature Communications (2020)
-
Live-cell super-resolved PAINT imaging of piconewton cellular traction forces
Nature Methods (2020)