Abstract
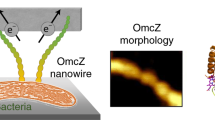
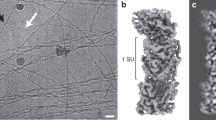
OmcZ nanowires produced by Geobacter species have high electron conductivity (>30 S cm−1). Of 111 cytochromes present in G. sulfurreducens, OmcZ is the only known nanowire-forming cytochrome essential for the formation of high-current-density biofilms that require long-distance (>10 µm) extracellular electron transport. However, the mechanisms underlying OmcZ nanowire assembly and high conductivity are unknown. Here we report a 3.5-Å-resolution cryogenic electron microscopy structure for OmcZ nanowires. Our structure reveals linear and closely stacked haems that may account for conductivity. Surface-exposed haems and charge interactions explain how OmcZ nanowires bind to diverse extracellular electron acceptors and how organization of nanowire network re-arranges in different biochemical environments. In vitro studies explain how G. sulfurreducens employ a serine protease to control the assembly of OmcZ monomers into nanowires. We find that both OmcZ and serine protease are widespread in environmentally important bacteria and archaea, thus establishing a prevalence of nanowire biogenesis across diverse species and environments.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
The key relevant datasets generated during and/or analysed during the current study are publicly available. Cryo-EM data were deposited with the Electron Microscopy Data Bank (ID EMD-23481) and with the Protein Data Bank (ID 7LQ5). Source data are provided with this paper.
References
Yalcin, S. E. & Malvankar, N. The blind men and the filament: understanding structures and functions of microbial nanowires. Curr. Opin. Chem. Biol. 59, 193–201 (2020).
Malvankar, N. S. et al. Tunable metallic-like conductivity in microbial nanowire networks. Nat. Nanotechnol. 6, 573–579 (2011).
Summers, Z. M. et al. Direct exchange of electrons within aggregates of an evolved syntrophic coculture of anaerobic bacteria. Science 330, 1413–1415 (2010).
Childers, S. E., Ciufo, S. & Lovley, D. R. Geobacter metallireducens accesses insoluble Fe(III) oxide by chemotaxis. Nature 416, 767–769 (2002).
Reguera, G. et al. Extracellular electron transfer via microbial nanowires. Nature 435, 1098–1101 (2005).
Malvankar, N. S. & Lovley, D. R. Microbial nanowires for bioenergy applications. Curr. Opin. Biotechnol. 27, 88–95 (2014).
Malvankar, N. S. & Lovley, D. R. Microbial nanowires: a new paradigm for biological electron transfer and bioelectronics. ChemSusChem 5, 1039–1046 (2012).
Wang, F. et al. Structure of microbial nanowires reveals stacked hemes that transport electrons over micrometers. Cell 177, 361–369 (2019).
Filman, D. J. et al. Cryo-EM reveals the structural basis of long-range electron transport in a cytochrome-based bacterial nanowire. Commun. Biol. 2, 219 (2019).
Yalcin, S. E. et al. Electric field stimulates production of highly conductive microbial OmcZ nanowires. Nat. Chem. Biol. 16, 1136–1142 (2020).
Ye, Y. et al. Dissecting the structural and conductive functions of nanowires in Geobacter sulfurreducens electroactive biofilms. mBio 13, e03822–03821 (2022).
Wang, F. et al. Cryo-EM structure of an extracellular Geobacter OmcE cytochrome filament reveals tetrahaem packing. Nat. Microbiol. 7, 1291–1300 (2022).
Gu, Y. et al. Structure of Geobacter pili reveals secretory rather than nanowire behavior. Nature 597, 430–434 (2021).
Nevin, K. P. et al. Anode biofilm transcriptomics reveals outer surface components essential for high density current production in Geobacter sulfurreducens fuel cells. PLoS ONE 4, e5628 (2009).
Yun, J., Malvankar, N. S., Ueki, T. & Lovley, D. R. Functional environmental proteomics: elucidating the role of a c-type cytochrome abundant during uranium bioremediation. ISME J. 10, 310–320 (2016).
Inoue, K. et al. Specific localization of the c-type cytochrome OmcZ at the anode surface in current-producing biofilms of Geobacter sulfurreducens. Environ. Microbiol. Rep. 3, 211–217 (2010).
Chadwick, G. L., Otero, F. J., Gralnick, J. A., Bond, D. R. & Orphan, V. J. NanoSIMS imaging reveals metabolic stratification within current-producing biofilms. Proc. Natl Acad. Sci. USA 116, 20716–20724 (2019).
Peng, L. & Zhang, Y. Cytochrome OmcZ is essential for the current generation by Geobacter sulfurreducens under low electrode potential. Electrochim. Acta 228, 447–452 (2017).
Richter, H. et al. Cyclic voltammetry of biofilms of wild type and mutant Geobacter sulfurreducens on fuel cell anodes indicates possible roles of OmcB, OmcZ, type IV pili, and protons in extracellular electron transfer. Energy Environ. Sci. 2, 506–516 (2009).
Gaffney, E. M. & Minteer, S. D. A silver assist for microbial fuel cell power. Science 373, 1308–1309 (2021).
Cao, B. et al. Silver nanoparticles boost charge-extraction efficiency in Shewanella microbial fuel cells. Science 373, 1336–1340 (2021).
Yi, H. et al. Selection of a variant of Geobacter sulfurreducens with enhanced capacity for current production in microbial fuel cells. Biosens. Bioelectron. 24, 3498–3503 (2009).
Malvankar, N. S., Tuominen, M. T. & Lovley, D. R. Biofilm conductivity is a decisive variable for high-current-density Geobacter sulfurreducens microbial fuel cells. Energy Environ. Sci. 5, 5790–5797 (2012).
Malvankar, N. S. et al. Electrical conductivity in a mixed-species biofilm. Appl. Environ. Microbiol. 78, 5967–5971 (2012).
Yalcin, S. E. & Malvankar, N. S. Seeing is believing: novel imaging methods help identify structure and function of Geobacter nanowires in electricity-producing biofilms in Roadmap on emerging concepts in the physical biology of bacterial biofilms: from surface sensing to community formation. Phys. Biol. 18, 051501 (2021).
O’Brien, J. P. & Malvankar, N. S. A simple and low-cost procedure for growing Geobacter sulfurreducens cell cultures and biofilms in bioelectrochemical systems. Curr. Protoc. Microbiol. 43, A.4K.1–A.4K.27 (2017).
Malvankar, N. S., Tuominen, M. T. & Lovley, D. R. Lack of cytochrome involvement in long-range electron transport through conductive biofilms and nanowires of Geobacter sulfurreducens. Energy Environ. Sci. 5, 8651–8659 (2012).
Malvankar, N. S., Rotello, V. M., Tuominen, M. T. & Lovley, D. R. Reply to ‘Measuring conductivity of living Geobacter sulfurreducens biofilms’. Nat. Nano 11, 913–914 (2016).
Leang, C., Malvankar, N. S., Franks, A. E., Nevin, K. P. & Lovley, D. R. Engineering Geobacter sulfurreducens to produce a highly cohesive conductive matrix with enhanced capacity for current production. Energy Environ. Sci. 6, 1901–1908 (2013).
Inoue, K. et al. Purification and characterization of omcZ, an outer-surface, octaheme c-type cytochrome essential for optimal current production by Geobacter sulfurreducens. Appl. Environ. Microbiol. 76, 3999–4007 (2010).
Dahl, P. J. et al. A 300-fold conductivity increase in microbial cytochrome nanowires due to temperature-induced restructuring of hydrogen bonding networks. Sci. Adv. 8, eabm7193 (2022).
Tan, Y. et al. Synthetic biological protein nanowires with high conductivity. Small 12, 4481–4485 (2016).
Qian, X. et al. Biochemical characterization of purified OmcS, a c-type cytochrome required for insoluble Fe(III) reduction in Geobacter sulfurreducens. Biochim. Biophys. Acta 1807, 404–412 (2011).
Thirumurthy, M. A. & Jones, A. K. Geobacter cytochrome OmcZs binds riboflavin: implications for extracellular electron transfer. Nanotechnology 31, 124001 (2020).
Okamoto, A. et al. Uptake of self-secreted flavins as bound cofactors for extracellular electron transfer in Geobacter species. Energy Environ. Sci. 7, 1357–1361 (2014).
Lovely, D. R. et al. Geobacter: the microbe electric’s physiology, ecology, and practical applications. Adv. Microb. Physiol. 59, 1–100 (2011).
Fukushima, T. et al. The molecular basis for binding of an electron transfer protein to a metal oxide surface. J. Am. Chem. Soc. 139, 12647–12654 (2017).
Shapiro, D. et al. Protein nanowires with tunable functionality and programmable self-assembly using sequence-controlled synthesis. Nat. Commun. 13, 829 (2021).
Kai, A. et al. Proteolytic maturation of the outer membrane c-type cytochrome OmcZ by a subtilisin-like serine protease is essential for optimal current production by Geobacter sulfurreducens. Appl. Environ. Microbiol. 87, e02617–e02620 (2021).
Jumper, J. et al. Highly accurate protein structure prediction with AlphaFold. Nature 596, 583–589 (2021).
Tauzin, A. S. et al. Molecular dissection of xyloglucan recognition in a prominent human gut symbiont. mBio 7, e02134–02115 (2016).
Lovley, D. R. Microbial nanowires. Curr. Biol. 32, R110–R112 (2022).
Chan, C. H., Levar, C. E., Jiménez-Otero, F. & Bond, D. R. Genome scale mutational analysis of Geobacter sulfurreducens reveals distinct molecular mechanisms for respiration and sensing of poised electrodes versus Fe(III) oxides. J. Bacteriol. 199, e00340–00317 (2017).
Chadwick, G. L. et al. Comparative genomics reveals electron transfer and syntrophic mechanisms differentiating methanotrophic and methanogenic archaea. PLoS Biol. 20, e3001508 (2022).
Malvankar, N. S., King, G. M. & Lovley, D. R. Centimeter-long electron transport in marine sediments via conductive minerals. ISME J. 9, 527–531 (2014).
Wegener, G., Krukenberg, V., Riedel, D., Tegetmeyer, H. E. & Boetius, A. Intercellular wiring enables electron transfer between methanotrophic archaea and bacteria. Nature 526, 587–590 (2015).
Scheller, S., Yu, H., Chadwick, G. L., McGlynn, S. E. & Orphan, V. J. Artificial electron acceptors decouple archaeal methane oxidation from sulfate reduction. Science 351, 703–707 (2016).
Shipps, C. et al. Intrinsic electronic conductivity of individual atomically-resolved amyloid crystals reveals micrometer-long hole hopping via tyrosines. Proc. Natl Acad. Sci. USA 118, e2014139118 (2021).
Malvankar, N. S., Mester, T., Tuominen, M. T. & Lovley, D. R. Supercapacitors based on c-type cytochromes using conductive nanostructured networks of living bacteria. ChemPhysChem 13, 463–468 (2012).
Neu, J. et al. Microbial biofilms as living photoconductors due to ultrafast electron transfer in cytochrome OmcS nanowires. Nat. Commun. 13, 5150 (2022).
Alexeyev, M. F. The pKNOCK series of broad-host-range mobilizable suicide vectors for gene knockout and targeted DNA insertion into the chromosome of Gram-negative bacteria. Biotechniques 26, 824–828 (1999).
Zivanov, J. et al. New tools for automated high-resolution cryo-EM structure determination in RELION-3. eLife 7, e42166 (2018).
Zhang, K. Gctf: real-time CTF determination and correction. J. Struct. Biol. 193, 1–12 (2016).
Pettersen, E. F. et al. UCSF Chimera—a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004).
Glaser, F. et al. ConSurf: identification of functional regions in proteins by surface-mapping of phylogenetic information. Bioinformatics 19, 163–164 (2003).
Goddard, T. D. et al. UCSF ChimeraX: meeting modern challenges in visualization and analysis. Protein Sci. 27, 14–25 (2018).
Case, D. A. et al. The Amber biomolecular simulation programs. J. Comput. Chem. 26, 1668–1688 (2005).
Case, D. A. et al. Amber20 (Univ. California, 2020); https://ambermd.org/CiteAmber.php
Hornak, V. et al. Comparison of multiple Amber force fields and development of improved protein backbone parameters. Proteins 65, 712–725 (2006).
Crespo, A. et al. Theoretical study of the truncated hemoglobin HbN: exploring the molecular basis of the NO detoxification mechanism. J. Am. Chem. Soc. 127, 4433–4444 (2005).
Henriques, J., Costa, P. J., Calhorda, M. J. & Machuqueiro, M. Charge parametrization of the DvH-c3 heme group: validation using constant-(pH,E) molecular dynamics simulations. J. Phys. Chem. B 117, 70–82 (2013).
Cruzeiro, V. W. D., Amaral, M. S. & Roitberg, A. E. Redox potential replica exchange molecular dynamics at constant pH in AMBER: implementation and validation. J. Chem. Phys. 149, 072338 (2018).
Cruzeiro, V. W. D., Feliciano, G. T. & Roitberg, A. E. Exploring coupled redox and pH processes with a force-field-based approach: applications to five different systems. J. Am. Chem. Soc. 142, 3823–3835 (2020).
Jorgensen, W. L., Chandrasekhar, J., Madura, J. D., Impey, R. W. & Klein, M. L. Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 79, 926–935 (1983).
Joung, I. S. & Cheatham, T. E. III Determination of alkali and halide monovalent ion parameters for use in explicitly solvated biomolecular simulations. J. Phys. Chem. B 112, 9020–9041 (2008).
Darden, T., York, D. & Pedersen, L. Particle mesh Ewald: An N⋅log(N) method for Ewald sums in large systems. J. Chem. Phys. 98, 10089–10092 (1993).
Ryckaert, J.-P., Ciccotti, G. & Berendsen, H. J. Numerical integration of the cartesian equations of motion of a system with constraints: molecular dynamics of n-alkanes. J. Comput. Phys. 23, 327–341 (1977).
Miyamoto, S. & Kollman, P. A. Settle: an analytical version of the SHAKE and RATTLE algorithm for rigid water models. J. Comput. Chem. 13, 952–962 (1992).
Gotz, A. W. et al. Routine microsecond molecular dynamics simulations with AMBER on GPUs. 1. Generalized born. J. Chem. Theory Comput. 8, 1542–1555 (2012).
Beck, A. D. Density-functional thermochemistry. III. The role of exact exchange. J. Chem. Phys. 98, 5648–5646 (1993).
Stephens, P. J., Devlin, F. J., Chabalowski, C. F. & Frisch, M. J. Ab initio calculation of vibrational absorption and circular dichroism spectra using density functional force fields. J. Phys. Chem. 98, 11623–11627 (1994).
Weigend, F. & Ahlrichs, R. Balanced basis sets of split valence, triple zeta valence and quadruple zeta valence quality for H to Rn: design and assessment of accuracy. Phys. Chem. Chem. Phys. 7, 3297–3305 (2005).
Suo, B., Shen, K., Li, Z. & Liu, W. Performance of TD-DFT for excited states of open-shell transition metal compounds. J. Phys. Chem. A 121, 3929–3942 (2017).
Curutchet, C. & Mennucci, B. Toward a molecular scale interpretation of excitation energy transfer in solvated bichromophoric systems. J. Am. Chem. Soc. 127, 16733–16744 (2005).
Russo, V., Curutchet, C. & Mennucci, B. Towards a molecular scale interpretation of excitation energy transfer in solvated bichromophoric systems. II. The through-bond contribution. J. Phys. Chem. B 111, 853–863 (2007).
Frisch, M. J. et al. Gaussian 16 Rev. A.03 (Gaussian, 2016); https://gaussian.com/citation_a03/
Jurinovich, S., Cupellini, L., Guido, C. A. & Mennucci, B. EXAT: excitonic analysis tool. J. Comput. Chem. 39, 279–286 (2018).
Jurinovich, S., Pescitelli, G., Di Bari, L. & Mennucci, B. A TDDFT/MMPol/PCM model for the simulation of exciton-coupled circular dichroism spectra. Phys. Chem. Chem. Phys. 16, 16407–16418 (2014).
Jurinovich, S., Guido, C. A., Bruhn, T., Pescitelli, G. & Mennucci, B. The role of magnetic–electric coupling in exciton-coupled ECD spectra: the case of bis-phenanthrenes. Chem. Commun. 51, 10498–10501 (2015).
Graves, A. B., Graves, M. T. & Liptak, M. D. Measurement of heme ruffling changes in MhuD using UV–vis spectroscopy. J. Phys. Chem. B 120, 3844–3853 (2016).
Sarkar, R., Boggio-Pasqua, M., Loos, P.-F. & Jacquemin, D. Benchmarking TD-DFT and wave function methods for oscillator strengths and excited-state dipole moments. J. Chem. Theory Comput. 17, 1117–1132 (2021).
Adamo, C. & Jacquemin, D. The calculations of excited-state properties with time-dependent density functional theory. Chem. Soc. Rev. 42, 845–856 (2013).
Londer, Y. Y., Pokkuluri, P. R., Tiede, D. M. & Schiffer, M. Production and preliminary characterization of a recombinant triheme cytochrome c7 from Geobacter sulfurreducens in Escherichia coli. Biochim. Biophys. Acta 1554, 202–211 (2002).
Skerra, A. & Schmidt, T. G. Use of the Strep-tag and streptavidin for detection and purification of recombinant proteins. Methods Enzymol. 326, 271–304 (2000).
Arslan, E., Schulz, H., Zufferey, R., Künzler, P. & Thöny-Meyer, L. Overproduction of the Bradyrhizobium japonicum c-type cytochrome subunits of the cbb3 oxidase in Escherichia coli. Biochem. Biophys. Res. Commun. 251, 744–747 (1998).
Siezen, R. J. & Leunissen, J. A. Subtilases: the superfamily of subtilisin‐like serine proteases. Protein Sci. 6, 501–523 (1997).
Tang, G. et al. EMAN2: an extensible image processing suite for electron microscopy. J. Struct. Biol. 157, 38–46 (2007).
Desfosses, A., Ciuffa, R., Gutsche, I. & Sachse, C. SPRING—an image processing package for single-particle based helical reconstruction from electron cryomicrographs. J. Struct. Biol. 185, 15–26 (2014).
Evans, R. et al. Protein complex prediction with AlphaFold-Multimer. Preprint at bioRxiv https://doi.org/10.1101/2021.10.04.463034 (2022).
Zemla, A. LGA: a method for finding 3D similarities in protein structures. Nucleic Acids Res. 31, 3370–3374 (2003).
Kumar, S. et al. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 35, 1547 (2018).
Edgar, R. C. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32, 1792–1797 (2004).
Letunic, I. & Bork, P. Interactive Tree Of Life (iTOL) v4: recent updates and new developments. Nucleic Acids Res. 47, W256–W259 (2019).
Acknowledgements
We thank E. Martz for help with structural analysis, C. Shipps for help with UV–vis spectroscopy of reduced OmcZ nanowires, and T. Pollard, M. Hochstrasser and B. Kazmierczak for helpful suggestions. This research was supported by the NSF CAREER award no. 1749662 (to N.S.M.), the NSF EAGER award no. 2038000 (to N.S.M.), the NSF-ANR award no. 2210473 (to N.S.M. and V.S.B.) and the National Institutes of Health Director’s New Innovator award (1DP2AI138259-01 to N.S.M. and R01GM141192 to K.G.). Research was sponsored by the Defense Advanced Research Project Agency Army Research Office and was accomplished under Cooperative Agreement Number W911NF-18-2-0100 (with N.S.M. and V.S.B.). High-resolution cryo-EM data collection was performed at the Case Western Reserve University Cryo-Electron Microscopy Core facility with the assistance of Kunpeng Li and Sudha Chakrapani.
Author information
Authors and Affiliations
Contributions
Y.G. purified OzpA, OmcZ50 and OmcZ nanowires, collected and analysed negative-stain and cryo-EM data to build the atomic model, performed biochemical analyses, UV–vis, CD spectroscopy, solubility test, nanowire alignment, in vitro digestion experiment of OmcZ nanowires, immune-gold labelling and power spectra analyses. M.J.G.-P. performed molecular dynamics and CD simulations. Y.L. constructed the expression vector for OmcZ50 in E. coli. Y.G. and F.A.S. optimized the culturing condition and purified OmcZ50 from E. coli. V.S. helped with OmcZ nanowire purification. F.A.S. and V.S. helped with model building. C.S. constructed the omcS:pk18 strain for OmcZ purification. F.G. performed the native mass spectrometry experiments and analysed the data under the supervision of K.G. V.S.B. guided computational studies. Y.G. and N.S.M. conceived and designed the project. N.S.M. supervised the work. Y.G. and N.S.M. wrote the paper with input from all the authors.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Microbiology thanks Thomas Boesen and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Illustration of cryo-EM map quality.
a–g, Density fitting of individual heme cofactors. Low iron density is due to the non-selection of coordinated histidine when rendering the figures at the given threshold of the density. i, Resolution distribution of the cryo-EM map. j, Representative density of sidechains. k, Map to map FSC. l, Map to model FSC. m, Real space correlation coefficient (RSCC) for each residue. n, Heme RSCC.
Extended Data Fig. 2 omcZ homologues show high sequence similarity.
omcZ homologues show up to 68% sequence similarity. In addition to all heme binding motifs, key residues are also highly conserved which we found to be critical for high conductivity. For example, a consecutive pair of histidines that brings T-stacked hemes closer is also highly conserved in all omcZ homologs. Thus significant sequence identities are present throughout the protein.
Extended Data Fig. 3 Circular Dichroism (CD) spectroscopy shows unique heme arrangements for OmcZ nanowires.
a, CD spectra OmcZ nanowire shows similar secondary structure at pH 10.5 and pH 7 buffer. b, CD spectra of OmcS and OmcZ nanowire hemes. Reduced spectrum show narrowing, and a red-shift of the peaks compared to the oxidized spectrum for c, OmcZ. d, OmcS nanowires as expected. e, The double positive peak feature is unique to the OmcZ nanowires, while the non-filamentous OmcZ30 fails to show a such feature. f, Simulated CD spectra for various sextets of hemes from OmcZ showing that the line shape is independent of six hemes included in the computations. Simulated CD spectra of g, OmcS (green) and h, OmcZ (pink), using only hemes and heme binding motifs show the peak features observed in the experimental data (black). The simulated spectra were uniformly shifted to longer wavelength by 0.06 eV (see methods for details) i, Resolving the contribution of individual hemes to the CD spectra for OmcS and OmcZ reveals that the double peak is a specific feature of hemes in OmcZ nanowires. Evolution of the CD spectra as adjacent hemes in OmcS (green) and OmcZ (pink) were sequentially added to the computations. The labels in each panel indicate which hemes were added for the spectra. The sequence of addition is from left-to-right in both rows.
Extended Data Fig. 4 OmcZ nanowires reduce diverse electron acceptors.
a, Buffer Control: OmcZ nanowire spectra do not change after 1 hour upon addition of degassed deionized bis-tris buffer (20 mM, pH=7.2) used for all substrate reduction experiments. b, OmcZ nanowires could not be fully reduced by DTT but could be reduced by Sodium dithionite. c, OmcZ cannot reduce riboflavin.
Extended Data Fig. 5 OmcZ nanowires are robust electronic biomaterials that function under extreme environments.
Negative staining TEM images of OmcZ nanowires treated with following protein denaturants. a, No denaturant. b, 6 M urea, c, 5 M GuHCl, d, Phosphoric acid at low pH (pH = 1.6), and e, boiled in 2% SDS. All scale bars: 100 nm. f, OmcZ nanowires are more stable than OmcS at low pH (pH = 1.6) as evident by retention of its red colour due to hemes. OmcZ nanowires also retained their red colour characteristic of haems under all other denaturants. g, Solution CD spectra and h, UV-vis spectra show the stability of protein and electronic structure of OmcZ nanowires in denaturants.
Extended Data Fig. 6 In vitro Assembly of OmcZ nanowires from native OmcZ50.
a, TEM image of native G. sulfurreducens OmcZ50 showing lack of nanowire assembly. b, Western immunoblot using OmcZ antibody showing OzpA digesting OmcZ50 into OmcZ30. c, Digested OmcZ30. self-assembles into nanowires in vitro. d, SDS-PAGE gel of purified OzpA showing three major bands: premature protein, mature protein, and the cleaved inhibitor domain (I9). e, Power spectrum of the reconstituted OmcZ nanowires shows the same helical parameters as cell-produced OmcZ nanowires. f, 2D average of reconstituted OmcZ nanowires. g, CD spectra of reconstituted and native OmcZ nanowires is similar. Immuno-gold labeling of reconstituted OmcZ nanowires using h, No-primary antibody, i, antibody for OmcZ30 and j, antibody for OmcZ50. Scale bars, a, 100 nm. c, 50 nm. c, 2 nm. h, 100 nm, i, 50 nm, j, 100 nm.
Extended Data Fig. 7 Native mass spectrometry of OmcZ50 purified from E. coli shows molecular weight and heme groups consistent with native OmcZ50.
a, Coomassie and Heme staining gel of OmcZ50 purified from E. coli. b, Mass spectrum of OmcZ purified from G. sulfurreducens. Detected mass corresponds to the OmcZ monomer covalently bound to eight hemes. c-d, Mass spectrum of OmcZ50 purified from E. coli. Detected proteins exist as monomer and homodimer. Mass analysis shows that OmcZ50 monomer is bound covalently to primarily c, eight haems but sometimes also to d, six hemes.
Extended Data Fig. 8 OmcZ nanowire assembly could be environmentally controlled.
a, omcZ operon genes with intergenic region in G. sulfurreducens. b, RT-PCR shows that omcZ and ozpA are co-transcribed. Lanes 1: genomic DNA (gDNA), 2: cDNA, 3: cDNA control. c, Predicted RNA structure of the intergenic region of mRNA using ViennaRNA suite. Color code shows the probability of each base or base pair’s placement in the secondary structure.
Extended Data Fig. 9 OmcZ operon is found in diverse microbes.
Phylogenetic tree derived from OmcZ amino acid sequence alignments using homology cut-off score of 100 bit. Light blue circles represent bootstrap values.
Supplementary information
Supplementary Information
Supplementary Tables 1–3 and Fig. 1.
Supplementary Video 1
OzpA cleaves OmcZ50 into OmcZ30 that self-assembles into nanowires.
Source data
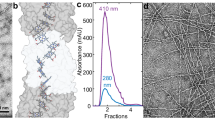
Source Data Fig. 1
Raw data for size exclusion chromatography.
Source Data Fig. 3
Raw data for iron reduction measurements.
Source Data Extended Data Fig. 1
Raw data for cryo-EM structural analysis.
Source Data Extended Data Fig. 3
Raw data for CD measurements and modelling.
Source Data Extended Data Fig. 4
Raw data for reduction measurements of electron acceptors.
Source Data Extended Data Fig. 5
Raw data for CD and UV–vis measurements.
Source Data Extended Data Fig. 6
Raw data for CD measurements.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Gu, Y., Guberman-Pfeffer, M.J., Srikanth, V. et al. Structure of Geobacter cytochrome OmcZ identifies mechanism of nanowire assembly and conductivity. Nat Microbiol 8, 284–298 (2023). https://doi.org/10.1038/s41564-022-01315-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41564-022-01315-5
This article is cited by
-
Widespread extracellular electron transfer pathways for charging microbial cytochrome OmcS nanowires via periplasmic cytochromes PpcABCDE
Nature Communications (2024)
-
Mechanisms of extracellular electron transfer in anaerobic methanotrophic archaea
Nature Communications (2024)
-
Multi-heme cytochrome-mediated extracellular electron transfer by the anaerobic methanotroph ‘Candidatus Methanoperedens nitroreducens’
Nature Communications (2023)