Abstract
So far, only members of the bacterial phyla Proteobacteria and Verrucomicrobia are known to grow methanotrophically under aerobic conditions. Here we report that this metabolic trait is also observed within the Actinobacteria. We enriched and cultivated a methanotrophic Mycobacterium from an extremely acidic biofilm growing on a cave wall at a gaseous chemocline interface between volcanic gases and the Earth’s atmosphere. This Mycobacterium, for which we propose the name Candidatus Mycobacterium methanotrophicum, is closely related to well-known obligate pathogens such as M. tuberculosis and M. leprae. Genomic and proteomic analyses revealed that Candidatus M. methanotrophicum expresses a full suite of enzymes required for aerobic growth on methane, including a soluble methane monooxygenase that catalyses the hydroxylation of methane to methanol and enzymes involved in formaldehyde fixation via the ribulose monophosphate pathway. Growth experiments combined with stable isotope probing using 13C-labelled methane confirmed that Candidatus M. methanotrophicum can grow on methane as a sole carbon and energy source. A broader survey based on 16S metabarcoding suggests that species closely related to Candidatus M. methanotrophicum may be abundant in low-pH, high-methane environments.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Data availability
Raw data from the proteomic analysis of the Sulfur Cave biofilm, annotation files and supplementary data can be found in a Zenodo repository72. Illumina 16S rRNA gene amplicon data from the Sulfur Cave biofilm are associated with the NCBI BioProject PRJNA675490. Illumina reads from the sequencing of the two metagenomes from the cave and one from the culture as well as the MAGs M. MAGs 1, 2 and 3 are deposited at ENA (PRJEB45004) with the accession numbers ERR10036468, ERR10036469, ERR10036470, ERS6581338, ERS6581340 and ERS6581341, respectively and accession CAJUXY010000000. The 16S rRNA gene sequence obtained from the Candidatus M. methanotrophicum culture is available at GenBank (MW243585). The Illumina and Nanopore reads from this culture as well as the resulting whole-genome assembly are available at NCBI under BioProject PRJNA837300.
References
Martins, Z. et al. Earth as a tool for astrobiology—a European perspective. Space Sci. Rev. 209, 43–81 (2017).
Vaselli, O. et al. A geochemical traverse across the Eastern Carpathians (Romania): constraints on the origin and evolution of the mineral water and gas discharges. Chem. Geol. 182, 637–654 (2002).
Frunzeti, N., Baciu, C., Etiope, G. & Pfanz, H. Geogenic emission of methane and carbon dioxide at Beciu mud volcano, (Berca-Arbǎnaşi hydrocarbon-bearing structure, Eastern Carpathians, Romania). Carpathian J. Earth Environ. Sci. 7, 159–166 (2012).
Althaus, T., Niedermann, S. & Erzinger, J. Noble gas studies of fluids and gas exhalations in the East Carpathians, Romania. Chem. Erde Geochem. 60, 189–207 (2000).
Sarbu, S. M. et al. Sulfur Cave (Romania), an extreme environment with microbial mats in a CO2–H2s/O2 gas chemocline dominated by mycobacteria. Int. J. Speleol. 47, 173–187 (2018).
Jones, D. S., Schaperdoth, I. & Macalady, J. L. Metagenomic evidence for sulfide oxidation in extremely acidic cave biofilms. Geomicrobiol. J. 31, 194–204 (2014).
Cosma, C. L., Sherman, D. R. & Ramakrishnan, L. The secret lives of the pathogenic mycobacteria. Annu. Rev. Microbiol. 57, 641–676 (2003).
Cook, G. M. et al. Physiology of Mycobacteria. Advances in Microbial Physiology 81–182. (2009).
Walsh, C. M., Gebert, M. J., Delgado-Baquerizo, M., Maestre, F. T. & Fierer, N.A global survey of mycobacterial diversity in soil. Appl. Environ. Microbiol. https://doi.org/10.1128/aem.01180-19 (2019).
Khan, A. & Sarkar, D. Nitrate reduction pathways in mycobacteria and their implications during latency. Microbiology 158, 301–307 (2012).
Brezna, B., Khan, A. A. & Cerniglia, C. E. Molecular characterization of dioxygenases from polycyclic aromatic hydrocarbon-degrading Mycobacterium spp. FEMS Microbiol. Lett. 223, 177–183 (2003).
Cook, G. M., Hards, K., Vilchèze, C., Hartman, T. & Berney, M. Energetics of respiration and oxidative phosphorylation in mycobacteria. Microbiol. Spectr. https://doi.org/10.1128/microbiolspec.mgm2-0015-2013 (2014).
Guerrero-Cruz, S. et al. Methanotrophs: discoveries, environmental relevance, and a perspective on current and future applications. Front. Microbiol. https://doi.org/10.3389/fmicb.2021.678057 (2021).
Gupta, R. S., Lo, B. & Son, J. Phylogenomics and comparative genomic studies robustly support division of the genus Mycobacterium into an emended genus Mycobacterium and four novel genera. Front. Microbiol. https://doi.org/10.3389/fmicb.2018.00067 (2018).
Colby, J., Stirling, D. I. & Dalton, H. The soluble methane mono-oxygenase of Methylococcus capsulatus (Bath). Its ability to oxygenate n-alkanes, n-alkenes, ethers, and alicyclic, aromatic and heterocyclic compounds. Biochem. J. 165, 395–402 (1977).
Stainthorpe, A., Lees, V., Salmond, G. P., Dalton, H. & Murrell, J. The methane monooxygenase gene cluster of Methylococcus capsulatus (Bath). Gene 91, 27–34 (1990).
Cardy, D. L. N., Laidler, V., Salmond, G. P. C. & Murrell, J. C. The methane monooxygenase gene cluster of Methylosinus trichosporium: cloning and sequencing of the mmoc gene. Arch. Microbiol. 156, 477–483 (1991).
Chistoserdova, L., Chen, S.-W., Lapidus, A. & Lidstrom, M. E. Methylotrophy in Methylobacterium extorquens AM1 from a genomic point of view. J. Bacteriol. 185, 2980–2987 (2003).
Harms, N. & van Spanning, R. J. M. C1 metabolism in Paracoccus denitrificans: genetics of Paracoccus denitrificans. J. Bioenerg. Biomembr. 23, 187–210 (1991).
Norin, A., Piersma, S. R., Duine, J. A. & Jörnvall, H. Nicotinoprotein (NAD+-containing) alcohol dehydrogenase: structural relationships and functional interpretations. Cell. Mol. Life Sci. 60, 999–1006 (2003).
Schenkels, P. & Duine, J. A. Nicotinoprotein (NADH-containing) alcohol dehydrogenase from Rhodococcus erythropolis DSM 1069: an efficient catalyst for coenzyme-independent oxidation of a broad spectrum of alcohols and the interconversion of alcohols and aldehydes the EMBL accession number for the sequence reported in this paper is p81747. Microbiology 146, 775–785 (2000).
Haft, D. H. Bioinformatic evidence for a widely distributed, ribosomally produced electron carrier precursor, its maturation proteins, and its nicotinoprotein redox partners. BMC Genomics https://doi.org/10.1186/1471-2164-12-21 (2011).
Dubey, A. A., Wani, S. R. & Jain, V. Methylotrophy in mycobacteria: dissection of the methanol metabolism pathway in Mycobacterium smegmatis. J. Bacteriol. https://doi.org/10.1128/jb.00288-18 (2018).
de Vries, G. E., Arfman, N., Terpstra, P. & Dijkhuizen, L. Cloning, expression, and sequence analysis of the Bacillus methanolicus C1 methanol dehydrogenase gene. J. Bacteriol. 174, 5346–5353 (1992).
Bystrykh, L. V. et al. Electron microscopic analysis and structural characterization of novel NADP(H)-containing methanol: N, N′-dimethyl-4-nitrosoaniline oxidoreductases from the Gram-positive methylotrophic bacteria Amycolatopsis methanolica and Mycobacterium gastri MB19. J. Bacteriol. 175, 1814–1822 (1993).
Diab, F. Succinate-mediated catabolite repression control on the production of glycine betaine catabolic enzymes in Pseudomonas aeruginosa PAO1 under low and elevated salinities. Microbiology 152, 1395–1406 (2006).
Quayle, J. in Carbohydrate Metabolism (ed. Shukla, A. K.) 360–364 (Elsevier, 1966).
Quayle, J. R. Microbial assimilation of C1 compounds. Biochem. Soc. Trans. 8, 1–10 (1980).
Kato, N., Yurimoto, H. & Thauer, R. K. The physiological role of the ribulose monophosphate pathway in bacteria and archaea. Biosci. Biotechnol. Biochem. 70, 10–21 (2006).
Serafini, A., Pisu, D., Palù, G., Rodriguez, G. M. & Manganelli, R. The ESX-3 secretion system is necessary for iron and zinc homeostasis in mycobacterium tuberculosis. PLoS ONE 8, e78351 (2013).
Sani, M. et al. Direct visualization by cryo-EM of the mycobacterial capsular layer: a labile structure containing ESX-1-secreted proteins. PLoS Pathog. 6, e1000794 (2010).
Howell, D. G. & McDaniel, H. A. Fluorescent staining of mycobacteria in bovine tissues with auramine O dye—a comparative evaluation of a modified staining procedure. Proc. Annu. Meet. U. S. Anim. Health Assoc. 71, 500–508 (1967).
Reed, W. M. & Dugan, P. R. Isolation and characterization of the facultative methylotroph Mycobacterium ID-y. Microbiology 133, 1389–1395 (1987).
Kambara, H. et al. Environmental factors affecting the community of methane-oxidizing bacteria. Microbes Environ. 37, n/a (2022). 10.1264/jsme2.ME21074
Draper, P. The outer parts of the mycobacterial envelope as permeability barriers. Front. Biosci. 3, d1253–1261 (1998).
Santos, R., Fernandes, J., Fernandes, N., Oliveira, F. & Cadete, M. Mycobacterium parascrofulaceum in acidic hot springs in Yellowstone National Park. Appl. Environ. Microbiol. 73, 5071–5073 (2007).
Melkonian, C. et al. High biodiversity in a benzene-degrading nitrate-reducing culture is sustained by a few primary consumers. Commun. Biol. https://doi.org/10.1038/s42003-021-01948-y (2021).
Cavicchioli, R. et al. Scientists’ warning to humanity: microorganisms and climate change. Nat. Rev. Microbiol. 17, 569–586 (2019).
Pratt, C. & Tate, K. Mitigating methane: emerging technologies to combat climate change’s second leading contributor. Environ. Sci. Technol. 52, 6084–6097 (2018).
Phan, T. H. et al. EspH is a hypervirulence factor for mycobacterium marinum and essential for the secretion of the ESX-1 substrates EspE and EspF. PLoS Pathog. 14, e1007247 (2018).
Tyanova, S., Temu, T. & Cox, J. The MaxQuant computational platform for mass spectrometry-based shotgun proteomics. Nat. Protoc.11, 2301–2319 (2016).
Weisburg, W. G., Barns, S. M., Pelletier, D. A. & Lane, D. J. 16S ribosomal DNA amplification for phylogenetic study. J. Bacteriol. 173, 697–703 (1991).
Kozich, J. J., Westcott, S. L., Baxter, N. T., Highlander, S. K. & Schloss, P. D. Development of a dual-index sequencing strategy and curation pipeline for analyzing amplicon sequence data on the MiSeq illumina sequencing platform. Appl. Environ. Microbiol. 79, 5112–5120 (2013).
Iturbe-Espinoza, P. et al. Effects of DNA preservation solution and DNA extraction methods on microbial community profiling of soil. Folia Microbiol. https://doi.org/10.1007/s12223-021-00866-0 (2021).
Li, D., Liu, C.-M., Luo, R., Sadakane, K. & Lam, T.-W. MEGAHIT: an ultra-fast single-node solution for large and complex metagenomics assembly via Succinct de Bruijn graph. Bioinformatics 31, 1674–1676 (2015).
Seah, B. K. B. & Gruber-Vodicka, H. R. gbtools: interactive visualization of metagenome bins in R. Front. Microbiol. https://doi.org/10.3389/fmicb.2015.01451 (2015).
Seemann, T. Prokka: rapid prokaryotic genome annotation. Bioinformatics 30, 2068–2069 (2014).
Jensen, L. J. et al. eggNOG: automated construction and annotation of orthologous groups of genes. Nucleic Acids Res. 36, D250–D254 (2007).
Parks, D. H., Imelfort, M., Skennerton, C. T., Hugenholtz, P. & Tyson, G. W. CheckM: assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res. 25, 1043–1055 (2015).
Jiang, H., Lei, R., Ding, S.-W. & Zhu, S. Skewer: a fast and accurate adapter trimmer for next-generation sequencing paired-end reads. BMC Bioinformatics 15, 182 (2014).
Wick, R. R., Judd, L. M., Gorrie, C. L. & Holt, K. E. Unicycler: resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput. Biol. 13, e1005595 (2017).
Jumper, J. et al. Highly accurate protein structure prediction with AlphaFold. Nature 596, 583–589 (2021).
Mirdita, M. et al. ColabFold—making protein folding accessible to all. Nat Methods. 19, 679–682 (2022).
Evans, R. et al. Protein complex prediction with AlphaFold-Multimer. Preprint at bioRxiv https://doi.org/10.1101/2021.10.04.463034 (2021).
Pettersen, E. F. et al. UCSF ChimeraX: structure visualization for researchers, educators, and developers. Protein Sci. 30, 70–82 (2020).
Jain, C., Rodriguez-R, L. M., Phillippy, A. M., Konstantinidis, K. T. & Aluru, S. High throughput ANI analysis of 90k prokaryotic genomes reveals clear species boundaries. Nat. Commun. https://doi.org/10.1038/s41467-018-07641-9 (2018).
Li, W. & Godzik, A. Cd-hit: a fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics 22, 1658–1659 (2006).
R: a language and environment for statistical computing (R Foundation for Statistical Computing, 2020).
Asnicar, F. et al. Precise phylogenetic analysis of microbial isolates and genomes from metagenomes using PhyloPhlAn 3.0. Nat. Commun. https://doi.org/10.1038/s41467-020-16366-7 (2020).
Buchfink, B., Xie, C. & Huson, D. H. Fast and sensitive protein alignment using DIAMOND. Nat. Methods 12, 59–60 (2014).
Edgar, R. C. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Research 32, 1792–1797 (2004).
Capella-Gutierrez, S., Silla-Martinez, J. M. & Gabaldon, T. trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 25, 1972–1973 (2009).
Price, M. N., Dehal, P. S. & Arkin, A. P. FastTree 2—approximately maximum-likelihood trees for large alignments. PLoS ONE 5, e9490 (2010).
Stamatakis, A. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30, 1312–1313 (2014).
Galili, T. dendextend: an R package for visualizing, adjusting, and comparing trees of hierarchical clustering. Bioinformatics https://doi.org/10.1093/bioinformatics/btv428 (2015).
Katoh, K., Misawa, K., Kuma, K. & Miyata, T. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 30, 3059–3066 (2002).
Letunic, I. & Bork, P. Interactive tree of life (iTOL) v4: recent updates and new developments. Nucleic Acids Res. 47, W256–W259 (2019).
Yu, G. Using ggtree to visualize data on tree-like structures. Curr. Protoc. Bioinformatics https://doi.org/10.1002/cpbi.96 (2020).
Veraart, A. J. et al. Living apart together—bacterial volatiles influence methanotrophic growth and activity. ISME J. 12, 1163–1166 (2018).
Meyer, N. R., Fortney, J. L. & Dekas, A. E. NanoSIMS sample preparation decreases isotope enrichment: magnitude, variability and implications for single-cell rates of microbial activity. Environ. Microbiol. 23, 81–98 (2020).
Polerecky, L. et al. Look@NanoSIMS—a tool for the analysis of nanoSIMS data in environmental microbiology. Environ. Microbiol. 14, 1009–1023 (2012).
Guan, Q. A methanotrophic Mycobacterium dominates a cave microbial ecosystem. Zenodo https://zenodo.org/record/4767037 (2021).
Fei, Q. et al. Bioconversion of natural gas to liquid fuel: opportunities and challenges. Biotechnol. Adv. 32, 596–614 (2014).
Vorholt, J. Cofactor-dependent pathways of formaldehyde oxidation in methylotrophic bacteria. Arch. Microbiol. 178, 239–249 (2002).
Hanson, R. S. & Hanson, T. E. Methanotrophic bacteria. Microbiol. Rev. 60, 439–471 (1996).
Deb, C. et al. A novel in vitro multiple-stress dormancy model for mycobacterium tuberculosis generates a lipid-loaded, drug-tolerant, dormant pathogen. PLoS ONE 4, e6077 (2009).
Acknowledgements
The authors thank C. Murrell for valuable discussions, C. P. Antony for assistance in metagenomics, A. Grootemaat for TEM imaging, M. Kienhuis for technical support with nanoSIMS analysis, I. Grigoriev for support during fluorescence microsopy, and Z. Para and B. Hegyeli, the Romanian custodians, for facilitating the experiments in Sulfur Cave. Also, we thank Utrecht Sequencing Facility (useq.nl) for providing sequencing service and data. The nanoSIMS facility at Utrecht University was financed through a large infrastructure grant by the Netherlands Organisation for Scientific Research (NWO, grant no. 175.010.2009.011). C.M. was supported by the Dutch Research Council, as part of the MiCRop Consortium (NWO/OCW grant no. 024.004.014). A.P. and Q.G. are supported by a faculty baseline grant (BAS/1/1020-01-01) from KAUST to A.P. The authors also thank members of the Bioscience Core Laboratory in KAUST for providing assistance with the generation of raw genome sequence datasets. PIE research was funded by FONDECYT-CONCYTEC (216-2015-FONDECYT). This is publication number 7467 of the Netherlands Institute of Ecology (NIOO-KNAW).
Author information
Authors and Affiliations
Contributions
R.J.M.v.S., organizing ideas, writing, sampling, pathway reconstructions and physiology. Q.G., R.U., J.G., C.M.B. and A.P., metagenomics and proteomics. C.M., writing, pathway reconstructions, phylogenomics and bioinformatic analyses. L.P., writing, 13C-methane experiments, fluorescence and nanoSIMS imaging, data analysis and interpretation. J.-F.F., whole-genome assembly and annotation. E.J.F., fruitful discussions. B.W.B., Illumina sequencing and data processing. J.W.A., sampling and fruitful discussions. M.B. and R.U., technical support. P.I.E., DNA extractions. M.M.M.-F. and P.L.E.B., culturing and 13C-methane experiments. S.R.P. and R.U., mass spectrometry. C.M.B., mycobacterial physiology. N.N.v.d.W., TEM imaging. V.D.G., sampling and culturing. S.M.S., sampling, culturing and fruitful discussions. W.B., organizing ideas, writing, sampling and mycobacterial genetics.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Microbiology thanks Marina Khalyuzhnaya, William Inskeep and Florin Musatfor their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
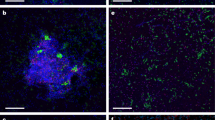
Extended Data Fig. 1 Sulfur Cave on the Puturosu Mountain (Stinky Mountain), Romania, from which Candidatus M. methanotrophicum was enriched.
(a) General overview of the cave. (b) Detailed images of the cave biofilms. The dashed line in panel a marks the stable gaseous chemocline between the volcanic gases (below the chemocline) and atmospheric air (above the chemocline). The cave walls below the chemocline are yellow due to sulphur deposition (marks 2, 5, 8), whereas no such depositions are present above the chemocline (marks 1, 3, 6). Biofilms on the cave walls are only present at the chemocline interface (mark 4). Mark 7 shows the bare cave wall after biofilm sampling.
Extended Data Fig. 2 Maps of sMMO gene clusters in known methanotrophs and selected members of the order Mycobacteriales.
(a) Gene clusters encoding sMMO in Methylosinu strichosporium OB3b, Methylococcus capsulatus Bath, which are known methanotrophs, and Candidatus M. methanotrophicum from this study. Abbreviations: hyp, gene encoding a hypothetical protein; B, mmoB; D, mmoD; Z, mmoZ; hyd, hydrogenase gene cluster; empty arrow, unknown ORF; r, hypothetical transcriptional regulator; PEP-ck, gene encoding phosphoenolpyruvate carboxykinase; SS, signal sensor of a two-component regulatory system; RR, response regulator of a two-component regulatory system. Each structural mmo gene is shown in distinct color, regulatory mmoR and groEL homologue mmoG genes are in red and orange, respectively. Candidatus M. methanotrophicum does not have mmoG in close proximity, but does have groEL genes elsewhere (see Extended Data Fig. 3). (b) Gene maps of genomic regions containing one or more of the mmoRXYBDCZ genes. The maps are shown for the species in Cluster 1 of the phylogenetic tree shown in Supplementary Fig. 4. Note that, in this figure, the direction of the arrows does not reflect the direction of gene transcription.
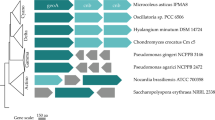
Extended Data Fig. 3 Maps of mft gene clusters in selected Mycobacterium species.
Abbreviations: M. MAG 1, Candidatus M. methanotrophicum; R, mftR; A, mftA; B, mftB; dac, D-aminocyclase gene; adh, alcohol dehydrogenase gene; hpr, hydroxypyruvate reductase gene; fdr, Ferredoxin-NAD(P)+ reductase gene; gmc, GMC-type oxidoreductase gene. adh from M. MAG 2 (ORF 0656) is closely related to the one from M. tuberculosis H37Rv (see also Supplementary Fig. 8).
Extended Data Fig. 4 Metabolic graph of the pentose phosphate pathway based on a corresponding KEGG map (map00030).
Shown are reactions of enzymes potentially encoded by the genes of Candidatus M. methanotrophicum (M. MAG 1, green), M. MAG 2 (red) and M. MAG 3 (purple) with the corresponding KEGG reaction IDs. Reactions with no color codes are present in all three M. MAGs. Orange circles represent intermediate compounds (corresponding KEGG compound IDs are shown in blue). KEGG reaction names: R05338: D-arabino-hex-3-ulose-6-phosphate formaldehyde-lyase (D-ribulose-5-phosphate-forming); R09780: D-arabino-hex-3-ulose-6-phosphate isomerase; R01741: D-Gluconate:(acceptor) 2-oxidoreductase; R02750: ATP:2-deoxy-D-ribose 5-phosphotransferase; R01051: ATP:D-ribose 5-phosphotransferase.
Extended Data Fig. 5 Number of unique proteins detected by LFQ intensity in the cave biofilm and in the enrichment culture of Candidatus M. methanotrophicum.
Only proteins from four prokaryotic species that remain in the enrichment culture are shown (see legend). Note the increasing relative contribution of proteins from Candidatus M. methanotrophicum in the enrichment culture compared to the cave biofilm.
Extended Data Fig. 6 Morphology and CO2 production of Candidatus M. methanotrophicum.
(a–b) Electron micrographs of Candidatus M. methanotrophicum (a) and M. smegmatis (a). Inset in panel a shows vesicles in the periplasm of Candidatus M. methanotrophicum at a higher magnification. Schematic representations of the micrographs in panels a’ and b’ show the cytosol (grey), the capsular layer (green), the plasma membrane (black line), the periplasm (yellow), the DNA (blue), the elucent areas (white), and the vesicles (red). (c–d) Evolution of CO2 during incubations of Candidatus M. methanotrophicum enrichment cultures with CH4. Incubations were conducted under aerobic (c) or anaerobic (d) conditions using either live cultures (green symbols) or a sterile NMS medium (black symbols). Different symbols correspond to replicate cultures (rep 1–3), and the values correspond to the total amount of CO2 per incubation bottle. Green lines show fits of the experimental data within the time interval of 4–22 days (c) and 0–22 days (d) with a linear model. For the replicate culture 1 incubated under aerobic conditions (rep 1), the slope of this linear model is significantly different from the corresponding negative slope of the linear model characterizing the removal of CH4 during the incubation (two-sided ANOVA, F = 6.79, p = 0.03; CH4 data shown in Fig. 3b). In contrast, the corresponding slopes are not significantly different for replicate cultures 2 (F = 0.0118, p = 0.92) and 3 (F = 3.38, p = 0.10). For both the aerobic and anaerobic incubations, the abiotic controls showed no significant variation in the CO2 amounts over time in comparison to the live cultures.
Extended Data Fig. 7 Additional nanoSIMS images of cells from Candidatus M. methanotrophicum enrichment cultures and cell biomass dynamics.
(a–d) Images of the 13C atom fraction, which is a measure of carbon assimilation from methane provided during the incubation. (e–h) Images of an overlay between the 12C14N ion counts intensity (blue), which is a proxy for biomass, and the 13C atom fraction (green). Cells shown in panels a–c and e–g were grown on 13C-labelled methane for 110 days, while cells in panels d and h were grown on unlabelled methane (control cells). Some cells in panels e–g appear blue because their 13C labeling is significantly lower compared to cells that appear cyan, although it was still significant compared to the control cells (see Fig. 4d). Note the filament in panels c and g, which belongs to a fungus from the genus Acidomyces. (i) Cell biomass as a function of time in two parallel subcultures of Candidatus M. methanotrophicum grown on CH4 as the sole carbon and energy source. One culture used 13C-labelled CH4 (13C atom fraction of 0.5), the other one used unlabelled CH4. Lines show the modelled biomass assuming an exponential growth with a doubling time of 94 days (solid line) and 98 days (dashed line) and a lag phase of 21 days. The decrease in the doubling time from 150–200 in the original culture to 94–98 days in these subcultures was possibly due to small changes in the culturing conditions combined with improved growth properties of Candidatus M. methanotrophicum. (j)13C atom fractions in the cells of Candidatus M. methanotrophicum grown on 13C-labelled CH4. Symbol shows the mean value, error bar corresponds to the standard deviation (calculated based on the measurement of N = 110 cells, where the cells were not treated by any post-incubation chemical procedure prior to the nanoSIMS analysis). Solid line shows the 13C atom fraction modelled based on the assumption that the 13C-labelled CH4 was the sole carbon and energy source and the growth characteristics were as shown in panel i.
Supplementary information
Supplementary Information
Supplemental materials, Tables 1–5, Figs. 1–12 and references.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
van Spanning, R.J.M., Guan, Q., Melkonian, C. et al. Methanotrophy by a Mycobacterium species that dominates a cave microbial ecosystem. Nat Microbiol 7, 2089–2100 (2022). https://doi.org/10.1038/s41564-022-01252-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41564-022-01252-3
This article is cited by
-
Targeting methanotrophs and isolation of a novel psychrophilic Methylobacter species from a terrestrial Arctic alkaline methane seep in Lagoon Pingo, Central Spitsbergen (78° N)
Antonie van Leeuwenhoek (2024)
-
Interkingdom interaction: the soil isopod Porcellio scaber stimulates the methane-driven bacterial and fungal interaction
ISME Communications (2023)
-
Simultaneous sulfide and methane oxidation by an extremophile
Nature Communications (2023)
-
Microbial methane cycling in a landfill on a decadal time scale
Nature Communications (2023)
-
Synergy effects of Methylomonas koyamae and Hyphomicrobium methylovorum under methanethiol stress
Applied Microbiology and Biotechnology (2023)